Beady-eyed bioassay can identify important nucleotides on the single molecule level
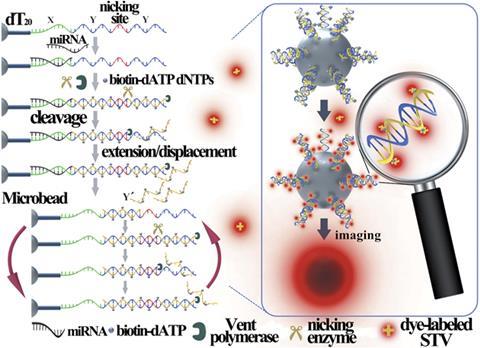
An elegant strategy for detecting minuscule quantities of microRNA using just a single functionalised microbead could be an important innovation for biomedical research and molecular diagnostics.1
Identifying biomolecules with vanishingly small copy numbers in cells is vital in deciphering the chemical blueprint for life. Homogeneous exponential amplification methods (such as the polymerase chain reaction), northern blotting, microarray detection and the isothermal exponential amplification reaction (EXPAR) are just some of the techniques that have undergone extensive tailoring to detect all manner of genetic material in cells as sensitively as possible. However, few approaches have even come close to reliably detecting nucleic acids at the single molecule level.
Zhengping Li and co-workers at Shaanxi Normal University, in China, have recently applied an isothermal EXPAR approach to unleash the potential of their new single microbead platform. With their design, the team succeeded in reliably detecting as little as 1 attomole of the microRNA let-7a, corresponding to just 3 copies of the molecule in a 5µL system.
A microRNA is a small, non-coding ribonucleic acid (RNA) molecule that carries out post-transcriptional regulation of gene expression. The human genome may encode hundreds of different microRNA molecules that play key roles in almost every biological process.2 Abnormal microRNA expression is linked to several human diseases, including cancer. ‘Since many human cancers often begin with cellular abnormalities in a small minority of cells within an organism, the detection of cell-to-cell variations of microRNA levels at the single cell level is extremely important in order to provide a deep insight into the function of microRNA, as well as the exact correlation between microRNA expression and cell function,’ explains Li.
The group’s system works by triggering an amplified response to the presence of a particular sequence of microRNA. The microbead is covered in identical strands (templates) of RNA and part of each is complementary to the target microRNA. When the target microRNA hybridises with one of these strands, a continuous extension, strand cleavage and strand displacement process begins. This incorporates nucleoside triphosphates – that will subsequently be tagged with fluorophores – all over the bead at an exponential rate. Therefore, when the target microRNA is present, a fluorescent halo is visible around the surface of the bead when viewed under a simple fluorescence microscope.
In traditional bead-based bioassays, thousands or millions of beads are used at a time and any fluorescence that develops in the vicinity of a target analyte will raise the average reading across all of the beads. Li points out that increased reliability and efficiency can be achieved by properly manipulating their single microbead. ‘It is clear that when the same numbers of fluorophores are gathered on a single bead rather than millions of beads, the fluorescence of the bead as well as the resulting detection sensitivity can be significantly enhanced.’
Chun-yang Zhang, based at Shandong Normal University, China, is an experienced researcher in the field of single molecule detection and imaging and sees tremendous promise in the strategy. ‘Traditional single molecule detection methods rely heavily on sophisticated and expensive instruments, and are only available in specialised labs,’ says Zhang. ‘This research enables detection on a single microbead, and might provide a simple and powerful tool for the early diagnosis of human diseases.’
‘The main innovation in this work is the utilisation of a single microbead as the carrier for signal amplification and signal enrichment,’ says Li. ‘We believe that, in the future, the single microbead-based sensing strategy can be easily extended to the detection of various genetic biomarkers, as well as protein biomarkers, with ultrahigh sensitivity.’
References
- X Zhang et al, Chem. Sci., 2015, DOI: 10.1039/c5sc02641e (This article is open access.)
I Bentwich et al, Nat. Genet., 2005, 37, 766 (DOI: 10.1038/ng1590)










No comments yet