Philip Ball explores the phenomenon of protein unfolding, and considers new techniques for keeping the egg unscrambled
Philip Ball explores the phenomenon of protein unfolding, and considers new techniques for keeping the egg unscrambled

Protein enzymes, like militant labourers, won’t work if the conditions aren’t right. Too hot, too cold, or too much pressure, and their delicately tuned molecular structures turn to useless spaghetti. You see the problem every time you cook an egg. Egg white is largely a protein called albumin, and heat changes it from a translucent, soluble form to a solid white mass. In cooked egg white, albumin’s exquisitely folded chain of amino acids is shaken apart and entangled with others. This loss of structure in a protein molecule is called denaturation.
The humble act of frying an egg thus serves as a metaphor for the fragility of all life on earth. For without protein-based enzymes to guide the body’s chemistry, life is impossible. You might call proteins Goldilocks molecules: the temperature (and pressure) has to be ’just right’. What’s more, they may unravel under extremes of pH or when exposed to certain small molecules called denaturants, such as urea. Why is this, and how (if at all) are different types of denaturation related?
Behind such questions is the profound issue of what makes an environment habitable. How can some organisms withstand, say, the superheated water that flows out of submarine volcanic vents? There are implications for technology too: for example, how can protein drugs be stored in ways that preserve their structure and function? Can we make proteins that remain active under extreme conditions? Why are some protein sequences - the ordering of amino acid groups in the protein chain - more resistant to heat than others?
And there are more pressing reasons to want to understand why proteins unfold and aggregate, since the aggregation of imperfectly folded forms underlies a host of neurodegenerative diseases, including Alzheimer’s, Parkinson’s, Huntington’s and Lou Gehrig’s. Here the aggregates are recalcitrant fibrils called amyloids that cause cell death in neurons. Understanding denaturation could eventually offer clues about how amyloid formation happens - and whether it can be avoided or even reversed.
Undone by cold
Proteins are spun on the cellular loom of the ribosome in an unfolded state, but the folding instructions are written into the protein sequence itself. Interactions among the amino acid groups, and with the water solvent, define a single most stable folded state. Key among these forces are hydrogen-bonding between amino acids and to the surrounding ’hydration shell’ of water, and the tendency of insoluble (hydrophobic) parts of the chain to clump together and get buried in the interior, away from water.
If a protein denatures, it can in principle find its way back to the ’native’, folded state. In fact this happens spontaneously all the time under the influence of thermal fluctuations. But there’s the danger that exposing hydrophobic groups along the chain may make denatured proteins stick together and aggregate into insoluble gunk, because the hydrophobic parts ’prefer’ to be in contact with each other rather than with water. Once that happens, it’s more or less irreversible - you can’t unboil an egg.
Whereas a huge amount of research has gone into understanding the protein folding process - the almost miraculous ability of a protein chain to find the most stable fold, often within just a millisecond - denaturation has been rather neglected, perhaps because it is less glamorous. The end result is not a beautifully efficient enzyme but a useless strand. But it’s increasingly clear that denaturation is not merely folding in reverse - it can occur in several ways.
Heat denaturation is a fairly simple process, the thermal motions just shake apart the weakly bound protein folds. ’It is driven by the increase of conformational entropy as the polypeptide chain partly unfolds,’ explains biophysicist Bertil Halle of Lund University in Sweden. But other types of denaturation have other causes.
Pressure-induced denaturation seems crudely to stem from the ’squeezing’ of solvent into the compact native state.
And for cold denaturation, the conventional view has been that the hydrophobic groups get more soluble in water (that is, the hydrophobic interaction is weakened) as the temperature is lowered. This explains why simple hydrophobic polymers, which collapse into compact states in water to minimise the contact with the solvent, swell in colder water. And traditionally that’s explained via an entropic driving force - water molecules close ranks and become more ordered around nonpolar hydrophobes, but this effect loses force as temperature drops.
But it now seems the truth is more subtle. Earlier this year, Halle looked at how the motions of water molecules surrounding proteins change as the temperature drops below the cold-denaturing transition.1 This happens below water’s freezing point, so Halle’s group suppressed ice formation by using microscopic water droplets suspended as an emulsion in oil. The tiny volume (about a picolitre, or 10-12 l) means that there is less chance of an ice crystal forming, allowing the water to be supercooled down to at least -35?C.
To probe water dynamics, the Lund team looked at the ’dispersion’ - basically the width - of the peaks in the nuclear magnetic resonance (NMR) signal from oxygen-17 in water enriched in this isotope. The dispersion reflects how quickly the water molecules in the hydration layer around a protein tumble. Halle and colleagues looked at four common ’model’ proteins, including apomyoglobin (the enzyme that binds oxygen in muscle tissue, with its active-site haem group removed), and b -lactoglobulin, a prominent component of milk whey.
Only the apomyoglobin denatured through cold alone, although b -lactoglobulin could be helped along the way by adding urea. But in neither case did this produce a fully unfolded state. Instead, the proteins remained relatively compact, as though water had penetrated within the native structure but not forced it apart. Halle thinks that, rather than being fully exposed to the solvent, a cold-denatured protein contains small clusters of water molecules interacting strongly with the peptide. ’The most important lesson from our work is that even though cold denaturation is driven by temperature-dependent hydration effects, it doesn’t involve the classical hydrophobic effect,’ he says.
Denaturants demystified
Indeed, there is now a growing view that all types of denaturation are intimately connected to changes in the way proteins are hydrated. In other words, the opening up of the native protein is not an intrinsic property of the polypeptide chain, but comes about in collaboration with the solvent. ’Protein stability is inseparable from protein-solvent interactions,’ says Halle.
That makes for a complex story, because water does unusual things when cooled or put under pressure. For example, it becomes less dense when cooled below 4?C, apparently as hydrogen-bonding between the molecules creates a more ’open’ structure akin to that in ice. Yet change in the structure of bulk water can’t be extrapolated to changes in the hydration layer, because the arrangements of water molecules can be rather different in the two cases.
All the same, this notion of bulk ’water structure’ as a kind of deus ex machina has mistakenly been invoked in the past to explain several unusual biomolecular behaviours, including the action of denaturants and the effect of salts on protein aggregation. One idea was that denaturants such as urea or guanidinium chloride (GdmCl) somehow perturb water’s bulk structure in a way that destabilises the folded protein, perhaps by altering the hydrophobic interactions that keep insoluble residues buried. This idea now looks increasingly flawed. Instead, we need to look at how denaturants affect the protein’s hydration shell.
For example, Jeremy England and Vijay Pande at Stanford University, US, working with Gilad Haran at the Weizmann Institute, Israel, used computer simulations to study the influence of urea and GdmCl on the hydrophobic interaction.2 As two hydrophobic surfaces come closer together in water, the water may suddenly be expelled from between them at a critical separation, and the surfaces are then pulled together by capillary forces. This ’dewetting transition’ has been proposed by David Chandler at the University of California at Berkeley, US, and his coworkers as an explanation of hydrophobic attraction in a folding protein.3
For two simple hydrophobic plates, England and colleagues found that urea gathers in the gap as clumps that sustain a liquid state there, suppressing the dewetting transition. This picture was anticipated in simulations by Bruce Berne (University of Columbia, US), Ruhong Zhou (IBM Watson Research Center, US) and their coworkers, who found that, for the model protein lysozyme, urea displaces water from the hydration shell and penetrates into the hydrophobic core, suppressing the native fold in favour of a swollen, rather disordered ’molten globule’.4
In England’s simulations GdmCl behaves differently, the molecules stick to the hydrophobic surfaces and give them less of a water-repelling veneer. Here the denaturant is acting somewhat like a surfactant, stabilising the interface between the surface and the solvent. But Berne and his colleagues have more recently found evidence that urea can act this way too - because it forms stronger dispersion forces than water with hydrophobic surfaces, it will accumulate there and mediate interactions with water through hydrogen bonds. In this way, Berne’s team found, urea can unravel a hydrophobic polymer in water.5 England suspects, however, that the stronger dispersion forces might be a consequence, rather than a cause, of urea’s attraction to hydrophobic surfaces.
Meanwhile, Brian Bennion and Valerie Daggett at the University of Washington, US, have found that urea not only alters hydrophobic interactions but also disrupts the hydration of hydrophilic parts of a protein, sticking there via hydrogen bonds.6 In effect, urea can usurp the hydrogen bonds that otherwise may help to bind the native state together. Walter Englander of the University of Pennsylvania, US, and his team have confirmed that urea denatures in this manner, while finding that GdmCl, rather surprisingly, doesn’t seem able to form comparable hydrogen bonds to the peptide chain.7
These ideas are still being debated, and many details have still to be ironed out, but the emerging picture is one in which denaturants exert their influence through direct interactions with the solute, not by restructuring the bulk solvent. And it seems that the same can also be said for the family of small molecules, such as glycerol, which have the opposite effect, stabilising a native protein against denaturation.
Some organisms use glycerol as a kind of antifreeze - but it may be that glycerol also acts here to stave off cold denaturation. A team led by Julio Fern?ndez at Columbia University, US, and colleagues tried to pull single molecules of ubiquitin apart by attaching one end of the chain to a surface and the other to the needle tip of an atomic-force microscope.8 Glycerol both stabilised the protein against this mechanical unfolding - more force was required to pull the chain out in this case - and also promoted hydrophobic collapse of the unfolded conformation. The researchers think that this stabilisation again happens through direct interactions with the protein, perhaps because the strongly polar glycerol hovering near the protein surface makes hydrophobic regions even less easily exposed.
Delicate balance

Whatever the case, it seems likely that there are several ways in which small molecules can destabilise proteins - making them look rather precarious. That’s increasingly the more general view, native proteins exist on an island (actually more of an isthmus) of stability hemmed in all around by denatured states. But are the denatured states formed by cold, heat, pressure and denaturants all parts of the same ’ocean’ on the map of temperature and pressure? If so, how are they related?
This global perspective has been explored in a simple lattice model of protein folding developed by Pablo Debenedetti and Frank Stillinger at Princeton University, US, and Peter Rossky at the University of Texas, US.9 They represent polypeptides as strings of hydrophobic and hydrophilic monomers surrounded by a two-dimensional square array of water molecules each with four hydrogen-bonding ’arms’. By imposing an entropic penalty and an energetic bonus for water-water bonds formed near hydrophobic monomers, the researchers found that these monomers effectively feel a hydrophobic attraction. In general, the polymers adopt either a relatively open conformation or a densely packed one with a hydrophobic core, which the researchers equate with the denatured and native states of proteins, respectively. But their ’cold-denatured’ states are not like the open-chain heat-denatured states, rather they are rearrangements of the compact state to break open the hydrophobic core and expose more of it to the solvent. This looks rather different to the ’solvent-expanded’ cold-denatured states inferred by Halle and colleagues; but that, Debenedetti admits, ’is simply a consequence of the crudeness of our model.’
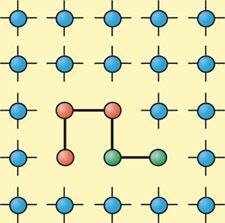
Nonetheless, the different denatured states merge smoothly with one other. As the pressure is raised, the heat- and cold-denaturing temperatures of proteins typically converge until they meet at a critical pressure, above which the folded state is never stable. ’There is a continuum of unfolded states, from cold-denatured states that expose hydrophobic residues maintaining compactness to heat-denatured states that unravel and truly unfold,’ says Debenedetti.
The next step, he adds, is to develop a more realistic model of protein structure - three-dimensional, and with genuine secondary structure such as helices and sheets rather than just globular compactness. He is now working on this with Rossky and postdoc Silvina Matysiak.
Designed for stability
Understanding protein denaturation could help us to avoid it, so that for example enzymes might be modified for use in biotechnology under non-physiological conditions. The replicase proteins used in the polymerase chain reaction (PCR) to amplify DNA sequences mustn’t unravel in the warmth needed to separate paired DNA strands for further replication. In this case the enzymes are taken from heat-loving (thermophilic) organisms, but such robust proteins are not always available from nature.
Debenedetti and colleagues’ lattice model offers clues to how a protein’s sequence influences stability of the native state. These chains, binary mixtures of hydrophilic and hydrophobic monomers, fall into two categories: those that have very stable compact structures and those that don’t. The former tend to contain long blocks of one monomer type, while the latter often have alternating monomer types. The researchers could perform a kind of directed evolution to improve the stability of some compact states, varying monomer type at random and selecting those sequences whose stability was improved. These were then used for the next round of mutation. The ’best’ mutations often, but not always, had elongated blocks of a single monomer type.
Such enhancement of native stability has already been demonstrated in real proteins. For example, Frances Arnold at the California Institute of Technology, US, and her coworkers have used in vitro directed evolution to improve the resistance to thermal denaturation in a variety of proteins.10 Typically, this is done by introducing random mutations into the gene encoding a protein, for example by using a version of PCR prone to making errors by inserting the wrong base, and then expressing the gene to make the mutant protein. The mutants are then screened for the desired properties, and the corresponding DNA sequences used for the next round of shuffling.
A crucial question is whether thermal stability can be improved without compromising the protein’s enzymatic activity. In nature there seems to be a trade-off, high thermal stability comes at the cost of relatively low catalytic activity, and vice versa. This may be because enzymes need to be somewhat flexible to carry out their catalytic tasks - but since thermal motions are smaller at lower temperatures, enzymes that work at increasingly low temperatures must be ever ’looser’ in structure, making them more easily disrupted by heat. Also, because reactions go faster at higher temperatures, thermophilic enzymes don’t need to be so finely tuned for high activity and so can afford to sacrifice some of that for the sake of stability.
’It’s much easier to be highly active or stable than to be highly active and stable,’ says Arnold, ’and since nature does not usually require high activity at low temperature and high thermostability, natural enzymes don’t usually optimise both properties.’ All the same, she doesn’t believe this is an intrinsic limitation. ’If you study natural proteins you only see a very limited subset of physically possible proteins, namely the biologically relevant ones,’ she says. ’Those reflect natural selection, not necessarily physical limitations.’
To prove the point, Arnold found that even cold-adapted proteins can be made stable against heat by directed evolution. For example, they converted a subtilisin enzyme from an Antarctic bacterium, which heat-denatures rather readily, into a thermophilic form with a denaturing temperature about 23?C higher.11
It’s hard to generalise about what works here. In some cases, it seems to be a matter of enhancing hydrophobic interactions in the protein core - in others, distant parts of the protein chain can hold other parts in place through hydrogen-bonding. In other cases, metal ions may form ’salt bridges’ linking together two parts of a chain. In short, it seems that more or less any of the forces that help to keep native states together can be commandeered to resist thermal denaturation. But if there are any general rules or principles, we’ve yet to discover them. And these effects will ultimately need to be understood not only in terms of how the protein interacts with itself but how it moulds and is moulded by its hydration shell.
Yet evidently nature can be ’improved’, proteins aren’t necessarily as stable as they could be. As we get to know more about why they unravel, we may find new ways to keep the egg unscrambled.
References
105 , 16928 (DOI: 10.1073/pnas.0808427105) et al, J. Chem. Phys., 2008, 128, 175102 (DOI: 10.1063/1.2909974)
10 F H Arnold et al, Trends Biochem. Sci., 2001, 26, 100 (DOI: 10.1016/S0968-0004(00)01755-2)
11 K Miyazaki et al, J. Mol. Biol., 2000, 297, 1015 (DOI: 10.1006/jmbi.2000.3612)






No comments yet