Can chemists make molecules that fold up as well as proteins? Rachel Brazil talks to the people trying to create foldamers
Molecules that fold are fundamental to life. ‘If you look at biology as a chemist, you can’t escape the conclusion that almost every complicated thing that biology does at the molecular level is carried out by a sequence-specific folded heteropolymer,’ says Sam Gellman from the University of Wisconsin–Madison in the US. Chemists have been trying to learn a few of these folding tricks from biology, but according to Jonathan Clayden from the University of Bristol in the UK, rather than just replicating these polymers, the aim now is ‘[to] do better than nature … with a bit of chemical ingenuity’. Using a wider spectrum of starting blocks he and others are creating molecules called foldamers that might one day beat biology at its own folding game.
The idea of synthesising molecules that could fold into secondary structures stems from work on protein folding carried out in the 1980s. ‘A key contribution was simulations from protein [modelling] specialist Ken Dill,’ says Gellman, an early adopter of the approach, who came up with the name ‘foldamer’.
Dill, now at Stony Brook University in New York state, US, had been working on protein folding and concluded that the process was driven by the juxtaposition of hydrophobic and hydrophilic amino acids in proteins. ‘Before that, the view had been that hydrogen bonding was the magic that dictated how proteins get their structure,’ says Dill. Work carried out by his collaborator Ron Zuckermann, then at pharma company Chiron, showed this was not the case. He used peptoids – made from poly-N-substituted glycines, which have side chains appended to the backbone nitrogen atom rather than the carbon. These molecules could adopt stable helices without the presence of hydrogen bonding, which convinced Dill and Zuckermann that folding was primarily due to the nature of amino acid side chains, with the backbone hydrogen bonding acting only as additional ‘glue’.
We walk in proteins’ footsteps, but we lag far behind
These ideas led Gellman to wonder what other molecules might be able to fold like peptides and he remembers questioning Dill after a conference talk, asking ‘If I could make a polystyrene with a hydrophobic styrene sub-unit and a hydrophilic styrene sub-unit, would they fold?’ The response was ‘Yes, I think so.’
For Dill and collaborator Zuckermann, the folding process is where life started and is responsible for the chemistry to biology transition. While the prevailing theory marks RNA as the first self-replicating molecule, Dill thinks that there must have been a stage before the ‘RNA world’ where molecules started folding, publishing his foldamer hypothesis in 2017.1 Dividing monomers into those with hydrophilic (polar) and those with hydrophobic side chains, he used a simple computer model to create chains where similar subunits were attracted to each other and found that even short chains can collapse into relatively compact structures.
‘There’s a natural elongation mechanism that is also selective and auto catalytic,’ Dill explains. This is because the collapsed structures expose what he calls ‘landing pads’ for catalysing other nascent polymers, ultimately creating primitive enzymes. ‘What it means is you’re going to have a whole ensemble of potential protein functions that are coming out of this soup, just naturally, because of the variability of hydrophobic–polar sequences themselves.’ For biology you ultimately needed information storage via DNA, but first you needed folding, Dill says.
So if biology is about folding, could chemists also harness this power? Gellman started trying in the 1990s, coming up with the name ‘foldamer’ for these types of synthetic molecules, typically 10–20 monomer units. ‘It turns out, you can’t do this with polystyrene because nobody knows how to make a polystyrene where you [can] control which monomer goes where, so a lot of this work has ended up focusing on polyamides,’ explains Gellman. He has focused on β-amino acids which have their amino group bonded to the β-carbon rather than the α as found in biology, but still fold into helices of various shapes, comparable to those found in proteins.
Others, such as supramolecular chemist Ivan Huc, from the University of Munich in Germany, have designed more exotic structures using aromatic oligoamides, and monomers bearing proteinogenic side chains that provide the folding impetus. Huc’s ‘apple peel’ helical capsule can be tuned in diameter according to monomer size, and specific attractive and repulsive interactions between the amide and the other functional groups can be substituted onto the aromatic rings. These foldamers can house a guest molecule in the resulting cavity.2 ‘These shapes are very trivial to obtain with aromatic amides and they are completely out of the reach of peptides or nucleotides,’ says Huc.
Designing foldamers is still a mostly trial and error process based on an understanding of local conformational preferences. Computational tools are gaining ground but aren’t as advanced as tools to model proteins and peptides. ‘We walk in their footsteps, but we lag far behind,’ says Huc.
Catalysis and drug discovery
‘One of the obvious dreams is to create catalytic versions [of foldamers],’ says Gellman, who recently took up this difficult challenge. In some cases, enzymes speed reactions up a million times by organising molecules within enclosed pockets. While Gellman cannot make this sort of tertiary structure yet, he did create a foldamer that allows two functional groups to be arranged in proximity to each other tethered to a helix.3 Gellman’s foldamer contained a and β amino acids, including β residues with five-membered rings, which stabilised the foldamer’s helical structure by constraining the backbone’s flexibility.
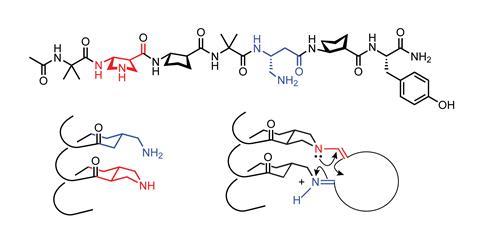
This was used to catalyse the formation of large macrocycles, which are useful as potential drugs but difficult to make as the two ends of long chain molecules need to be close together to react. Using a primary and secondary amine group each attached to a β residue, the foldamer is able to correctly position the ends and form a carbon–carbon bond via an aldol condensation, creating 12–22 carbon rings. Previous work had shown that such foldamer systems allowed similar reactions to proceed at least 100 times faster than using small molecule catalysts. The foldamer’s performance is still a long way from that of an enzyme though.
Gellman and others are also working on how foldamers could out-smart biology as drug molecules. ‘There is a whole host of peptides which sometimes are used as drugs but they break down [in the body] very fast,’ says Dimitri Dimitriou, chief executive of Swiss drug company Immupharma. ‘If you can effectively create a peptide analogue, which is stable, then [foldamers] have the potential to be as big as the monoclonal antibody industry – that’s the excitement from the commercial side.’ He is confident that within five years foldamer drugs will be on the market.
Gellman co-founded Longevity Biotech in 2010 to develop peptide drugs incorporating β-amino acids.4 ‘These peptides only have a quarter to a third of the β residue, but because they’re distributed along the backbone, proteolytic enzymes will cut [them] very slowly,’ he explains. The company call these helical foldamers ‘hybridtides’ and are trying to design hybridtide drugs that bind to G protein-coupled receptors (GPCRs), – transmembrane proteins that transmit signals inside a cell when stimulated by molecules outside. They are currently conducting a pre-clinical biomarker study for a Parkinson’s disease drug candidate.
During the coronavirus shutdown Gellman has continued to work on foldamers that may block the Sars-CoV-2 virus that causes Covid-19. The approach is based on work carried out in 2009 inspired by a drug for HIV–Aids.5 A 36-residue peptide, enfuvirtide, is effective in blocking the virus attaching itself to cells, but the drug has such a short half-life that patients needed to be injected twice a day. ‘We made variants that were 300-fold less susceptible to proteolysis [digestion] because of the a–β [backbone] and that’s what we’re trying to do with the coronavirus,’ says Gellman.
It’s a very complicated and difficult challenge – but this is what we are trying
Immupharma are also developing foldamer drugs alongside subsidiary company Ureka, based on the work of Giles Guichard at the University of Bordeaux in France. But their foldamers swap some amino acids for ureas, which have two amino groups joined by a carbonyl. ‘Oligourea is particularly good to form helices and those helices are similar to peptide helices … you have a good mixture of rigidity coming from the urea [backbone] and some flexibility coming from the sidechain groups, which can be substituted a little bit like an amino acid,’ explains Sebastien Goudreau, head of research at Ureka.
As proof-of-concept Ureka has started with glucagon-like peptide-1 (GLP-1), the 31-amino-acid hormone found in the pancreas that enhances the secretion of insulin and is used for the treatment of type 2 diabetes and the liver disease non-alcoholic-steatohepatitis. Their foldamer replaces four consecutive GLP-1 amino acids with three urea residues.6 ‘We have shown that it works and proved that it can extend the half-life dramatically [in mice],’ say Dimitriou. This could mean a dose would only be needed once a month and if resistant enough to digestive enzymes it might be able to be taken orally, although Dimitriou says they have not proven this yet.
Also on the radar are complex protein–protein interactions, traditionally considered undruggable. ‘It’s a very complicated and difficult challenge,’ says Huc. ‘But this is what we are trying.’ He has been designing foldamer molecules that can match a binding site in terms of their size, shape and proteinogenic side chains as far they can predict, but the final trick is to tether it to the protein. Using disulfide linkers, foldamers bearing different proteinogenic side chains were attached via a cysteine’s thiol side chain. Huc’s achiral foldamer will resonate between a left-handed and right-handed helix, but if it interacts with the protein surface, one version will become more favourable and predominate; this can be detected using circular dichroism spectroscopy.7 The sign of an interaction doesn’t mean tight binding, says Huc, but ‘from these interactions, I can design’.
Dynamic foldamers
Not only has nature created folded molecules, but also molecules that can change their shapes. For example, GPCRs will undergo conformational switching as they respond to hormones and the molecules that stimulate our senses of taste and smell. Clayden has been using foldamers to try and recreate the action of these receptors. ‘We’ve been designing molecules that have exactly the same sort of features – when they pick up a ligand for example, they change shape and as a result they transmit information through the structure of the molecule … that’s what we call dynamic foldamers.’
Unlike nature, Clayden starts with an achiral amino acid, α-aminoisobutyric acid (AIB). ‘You end up with a helix that can either be left- or right-handed and can actually inter-convert very rapidly between those,’ he says. The switching mechanism is provided by a large cyclic amino-borate group on the amine end of the foldamer. When a bulky chiral diol ligand is added it will form a boronate ester which then forms a methanol-bridge to the amine group. The steric bulk of the ligand forces the foldamer to switch to one helical sense.8 Clayden has shown these artificial receptors work when embedded in phospho-lipid vesicles.9 ‘[In] the long term we would like to get these things into real cells. We’ve done some very preliminary work,’ he says. These dynamic foldamers could lead to ‘smart’ drugs that could independently switch enzyme pathways on or off within cells depending on a specific stimulus.
Clayden has used the same approach to imitate our colour vision, which in nature relies on the GPCR receptor rhodopsin in the retinal rods. ‘Our molecule is an azobenzene chromophore and that’s attached to an AIB foldamer that changes shape when the azobenzene responds to light,’ he explains. In UV light the molecule switches to its cis conformation which induces a screw sense in the foldamer – making what Clayden calls a ‘conformational photo diode.’10 He envisions future smart chemical systems made from dynamic foldamers – for example, simply using different coloured lights to turn reactions on and off or switch from one enantiomeric product to another. ‘We’re currently working on a system that binds a catalyst, but releases it when it’s prompted to switch. That sort of idea could be used to release, for example, an enzyme inhibitor.’
Tertiary structure
Dill’s foldamer hypothesis for the early stages of life supposes a move from secondary folded structures to the proteins we have today, with their complex tertiary structures, combining helices and sheets made from defined peptide sequences. ‘The real power of biology in my view, and where I would love to see foldamers go, is hooking domains together,’ he says. But chemists are some way from this. ‘Most proteins are over 100 residues – that’s pretty hard for chemical synthesis,’ says Gellman.
Most labs are using solid-phase synthetic methods and starting to introduce automation but synthesising the relevant monomers isn’t trivial. ‘Small molecule synthesis is not nearly as advanced a field as it should be,’ says Gellman. For peptide chemistry, many of the starting blocks are commercially available but for foldamers that isn’t the case. ‘We can buy some of the β amino acids we need, but many of them, particularly when they have rings to constrain their local conformation, we can’t, and we don’t know how to make [them].’
Most proteins are over 100 residues – that’s pretty hard for chemical synthesis
Nevertheless chemists are attempting some simple tertiary structures. Several groups have produced foldamers that mimic the zinc finger domain (a protein motif that is able to coordination one or more zinc ions and binds a wide variety of biological molecules). Foldamers have also re-created the four-helix bundle motif, with hydrophobic residues buried in the core. Huc has even formed helical bundles in non-polar organic solvents – showing these structures can form in very different environments to nature.11
To create larger structures, Huc has suggested borrowing nature’s solution: ribosomes, the cell’s protein factory. ‘[My] long term dream is to hijack this machinery, and teach or modify the ribosome to produce [non-natural] chemical entities. This hasn’t been done yet and might not be so easy.’ Ribosomes are complexes of RNA and protein that are able to link amino acids together. They start with a messenger RNA (mRNA) template which base pairs with transfer RNA (tRNA) molecules that carry individual amino acids.
We need to think of other things that nature doesn’t do at all
Huc’s initial work with ribosomes in 2018 used novel RNA enzymes known as flexizymes, designed by Hiroaki Suga at the University of Tokyo in Japan, that are capable of attaching non-natural amino acids to tRNA. Huc was able to attach a dipeptide-appended aromatic helical foldamer. He then used an E. coli ribosome to synthesise a foldamer–peptide hybrid – the foldamer needed to unfold to get through the ribosome exit tunnel.12 While the ribosome is not forming bonds within the foldamer itself, it’s certainly a small step in that direction.
Going back 30 years the question was whether biological polymers and their ability to fold were unique. ‘Chemists have answered that: we can tell many different types of chemical backbones have a propensity to fold,’ says Huc. The question is now whether we can make increasingly complex large folded molecules – and what can we do with them. Nature has taught us some tricks, but chemists have a wider palette to work from. ‘[We need to] think of other things that nature doesn’t do at all,’ suggests Huc. ‘Perhaps the key developments will be in high temperature materials or micro-processors, who knows?’
Rachel Brazil is a science writer based in London, UK
References
1 E Guseva, R N Zuckermann and K A Dill, Proc. Natl Acad. Sci. USA, 2017, 114, E7460 (DOI: 10.1073/pnas.1620179114)
2 J Garric, J-M Léger and I Huc, Angew. Chem. Int. Ed., 2005, 44, 1954 (DOI: 10.1002/anie.200462898)
3 Z C Girvin, M K Andrews, X Liu, S H Gellman, Science, 2019, 366, 1528 (DOI: 10.1126/science.aax7344)
4 R Cheloha et al, Nat. Biotechnol., 2014, 32, 653 (DOI: 10.1038/nbt.2920)
5 S W Horne et al, Proc. Natl Acad. Sci. USA, 2009, 106, 14751 (DOI: 10.1073/pnas.0902663106)
6 J Fremaux et al, Nat Commun., 2019, 10, 924 (DOI: 10.1038/s41467-019-08793-y)
7 M Vallade et al, Bioconj. Chem., 2019, 30, 54 (DOI: 10.1021/acs.bioconjchem.8b00710)
8 R Brown et al, Nat. Chem., 2013, 5, 853 (DOI: 10.1038/nchem.1747)
9 F Lister et al, Nat. Chem., 2017, 9, 420 (DOI: 10.1038/nchem.2736)
10 D Mazzier et al, J. Am. Chem. Soc., 2016, 138, 8007 (DOI: 10.1021/jacs.6b04435)
11 S De et al, Nat. Chem., 2018, 10, 51 (DOI: 10.1038/nchem.2854)
12 J M Rogers et al, Nat. Chem., 2018, 10, 405 (DOI: 10.1038/s41557-018-0007-x)










No comments yet