We’ve known for a long time that different people respond to certain drugs to very different extents, but now cheap DNA testing could make these disparities a thing of the past, as Ian Le Guillou reports
Although best known today for his eponymous theorem, Pythagoras could also be considered a founding father of the burgeoning field of pharmacogenomics. Two and a half thousand years ago, the philosopher banned his followers from eating fava beans. Although his reasoning for this is unclear, the Mediterranean is today a hotspot for people with favism – a hereditary condition that makes consuming fava beans potentially fatal.
In some regions, as many as 30% of people have a mutation in the G6PD gene that impairs an enzyme important for red blood cell survival. When combined with an intake of fava beans, large numbers of the cells die, causing severe anaemia.
Today, pharmacogenomics is more concerned with the interaction between genes and drugs, rather than beans. For example, that same gene, G6PD, is essential for the body to process the anti-malarial drug primaquine. Munir Pirmohamed, a consultant physician and professor of pharmacology at the University of Liverpool, UK, remembers that was the first pharmacogenomic test that he used as a junior doctor in the early 1990s.
‘That brought back the pharmacological training I’d had as a medical student, and I was thinking this is something I can do more in the future,’ he recalls. ‘It was very clear from the beginning of my training that people respond differently to drugs. Some people respond well, some people don’t respond at all and some people respond adversely.’
Since then, Pirmohamed and the growing pharmacogenomics community have established over 400 gene–drug interactions. These are cases where common genetic variants can alter a person’s response to treatment, and changes in dosage or medication might be needed. Outside of a few narrow cases, however, these variants are not routinely tested for.
It was clear from the beginning of my training that people respond differently to drugs
Almost all of us have at least one genetic variant that affects how we process certain drugs. Although it is widely accepted by the medical community that genetics play a role in how patients respond to treatments, it is still rarely considered when prescribing. This is despite the potential for pharmacogenomics testing to significantly reduce side effects and improve treatment for patients. Healthcare staff could soon be confronted with these tests, however, as the evidence mounts for broader implementation and patients take matters into their own hands.
‘Many physicians and pharmacists agree that pharmacogenetics is important,’ says Jesse Swen, associate professor of pharmacogenetics and clinical pharmacist at Leiden University Medical Center in the Netherlands. ‘But they still consider it as something from the future – maybe in 10 years. They’re not aware that it’s so close to clinical application.
Potentially fatal
Many of the genes that have been identified in pharmacogenomics are linked to how we process drugs in our bodies. That includes the enzymes produced by the liver for metabolism or the proteins that transport the compounds to the kidneys for excretion. Mutations that affect these enzymes might never have any noticeable effect until the wrong medication is prescribed.
The reaction can be deadly. The DPYD gene is essential for metabolising the chemotherapy drugs 5-fluorouracil and capecitabine. Variants that reduce or eliminate the metabolism of these drugs cause an overexposure that is severely toxic. In 2020, NHS England issued an urgent policy statement recommending pre-treatment screening for DPYD variants with these drugs.
It has always surprised me that we don’t test for the gene variant
Unsurprisingly, it is the deadly gene–drug combinations that are among the few that have been prioritised for routine testing. Another example is the HIV treatment abacavir and variants of the gene HLA-B, which can cause the skin to blister and fall off. Previously, 5% of people taking the drug would have had this potentially fatal reaction, but with testing, this is now avoided.
Other variants have less severe immediate consequences but affect many more people. Mutations in the CYP2D6 gene can either increase or decrease the metabolic activity of its enzyme, which is responsible for metabolising a quarter of the drugs on the market. This includes several common antidepressants, which have a wide range of dosages for effective treatment without having toxic effects. Although these gene variants are unlikely to cause serious side effects, it takes more time to find the right dose by trial and error. ‘It has always surprised me that we don’t test for it,’ says Swen. ‘There’s absolutely benefit, in my opinion, for the patients, but it’s less obvious than with some of these very toxic drugs.’
CYP2D6 is also vital for metabolising codeine, so variants that eliminate the enzyme’s function prevent the drug from being converted into its active metabolite, morphine. ‘8% of people [in the UK] given codeine don’t get any relief of their pain. I feel that is fundamentally wrong for us to be able to do that,’ says Pirmohamed.
Extrapolating to the individual
Pharmacogenomics, also known as pharmacogenetics, is a subset of the wider field of precision medicine. The ultimate goal is to ensure that the right drug is given to the right patient at the right dose. Rather than trying to find the best treatment for a particular condition, pharmacogenomics focuses on the appropriate treatment for the person.
When I have a patient in my surgery, I have to extrapolate from the population to that individual
The shift to focusing on the individual being treated could better serve the challenges that doctors face in the clinic. ‘The problem with randomised controlled trials is that they are at a population level, and you look at the mean effect,’ says Pirmohamed. ‘But when I have a patient in my surgery coming to see me, I have to extrapolate from the population to that individual.’
Doctors and pharmacists can consider whether a patient has reduced kidney or liver function when adjusting the dosage of a drug. But patients whose genetics mean that they are completely lacking a functioning enzyme to metabolise the drug would be given the standard dose, unless testing is available. In time, pharmacogenomics could become just another tool available when making prescribing decisions. Pirmohamed suggests that it could even be possible to create algorithms that would account for a range of factors, including weight, sex, diet, liver and kidney function, other medication, and genetics.
We’re not testing for particular variants which may be occurring in certain ethnic groups
A key challenge for implementing pharmacogenomics is ensuring the tests are relevant to a broad range of patients amid the rapid pace of modern drug development. Pirmohamed led a clinical trial that showed in 2013 that testing for gene variants before prescribing warfarin improved the dosing for patients. However, warfarin use in wealthy countries is falling as its place is taken by newer anticoagulants that rely on different biochemical mechanisms. Warfarin is still commonly used in African countries, but results from a trial of mostly European people might not be applicable to other ethnicities. This is particularly true for Black African populations, which are the most genetically diverse group on Earth.
The tests that the NHS uses for the gene DPYD before prescribing certain chemotherapy drugs look for four variants that were largely identified in European ancestry populations. ‘We’re not testing for particular variants which may be occurring in certain ethnic groups,’ says Pirmohamed. He is now working to identify new variants in people with non-European ancestry to expand the testing panel. ‘Our population in the UK is diverse and I would hope that what we do over here has an impact in some way in other countries so that they can use the findings relevant to their population and incorporate them into their health care,’ he adds.
Panel testing
Rather than testing for gene variants one by one just before being prescribed a particular drug, it may be more efficient to examine a whole suite together in advance. While a particular variant might be uncommon in the population, almost all of us have at least one gene that could affect how we are prescribed medication.
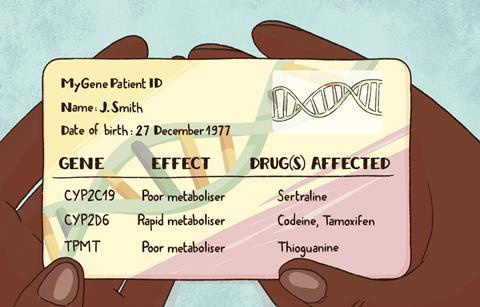
‘We did a study based on prescription data in the Netherlands from 2016. If we had known the pharmacogenetic profile of all Dutch citizens, then we would have adjusted almost 200,000 first prescriptions that year,’ says Swen. Out of a population of 17 million, this would only initially benefit a small proportion but the ongoing improvements accumulating each year would likely be significant.
Swen is testing this approach in a clinical trial across seven European countries with almost 7000 participants. Half of the patients in the trial were given a credit card-sized card with the results from a panel of pharmacogenomic markers that would be relevant to over 40 drugs. As the keepers of their own information, they could give it to their doctor when necessary, avoiding the need for a secure, centralised database that was easily accessible by all healthcare providers.
He suggests that if successfully scaled up, the panel test could be done for less than €100 (£85) – only slightly more expensive than a single test. Given that people who start a new medication are likely to need other treatments within the next few years, this could prove more cost-effective and reduce the waiting time to get the results before each new prescription.
Public acceptance
For most people, DNA testing is generally associated with police investigations, tracing ancestry or predicting the risk of inherited diseases. It is not information that people are comfortable with handing over to national databases. So helping people to accept the idea of genetics being part of routine testing could require careful handling.
‘You do need to explain that a pharmacogenetic profile is different from your risk for Alzheimer’s or other diseases; you have to explain that it’s more like measuring your kidney or liver function,’ says Swen.
Studies carried out by Swen and colleagues found that once people learn about pharmacogenomics, most would like to be profiled before starting a new drug and only 8% were opposed. This willingness was apparent in running the clinical trial, he says. ‘Recruitment overall with the project went exceptionally well for such a large project. People are interested in this topic when they are given the opportunity to participate.’
Many physicians aren’t really trained to know what to do
Although pharmacogenomics tests are not yet widely known about, some patients are already seeking them if they are concerned about their response to treatment. ‘Many [people] are actually searching on the internet and finding out, particularly if they had problems with their drugs. I get emails very regularly saying “I think I’ve got a pharmacogenomic problem, can you tell me where I can get this test done, because I’ve had problems with these drugs,”’ says Pirmohamed.
The gap in provision by healthcare systems has led to new avenues for the public to get tests. The direct-to-consumer genetics testing company 23andMe has been offering pharmacogenomic reports to the public since 2019, after receiving approval from the US Food and Drug Administration (FDA).
‘The information we provide them is really carefully considered by both us and the FDA. The main message is that we would like people to share this information with their healthcare provider,’ says Alison Chubb, a senior product scientist at 23andMe.
The company currently offers reports for three genes that affect drug metabolism. For some variants, it has also received approval to include information about the specific drugs that are affected, based on published guidelines.
‘Many physicians aren’t really trained to know what to do with [pharmacogenomics]. Many don’t know that the guidelines actually exist, and we feel like educating consumers about this information can help that information trickle up into the hands of the providers,’ says Chubb.
Rolling out more widely
The issue of education and training is brought up frequently in relation to pharmacogenomics. ‘There is a need to increase the understanding and the knowledge of pharmacogenetics in the clinical community as a whole, which includes not only doctors, but pharmacists and nurses,’ says Pirmohamed.
Managing a gene–drug interaction is similar to managing a drug–drug interaction
Pharmacists, in particular, could have an important role to play, as they are often the last healthcare professional that a patient will see before starting a new drug. Cheek swab tests could even be offered as part of a medication review, with recommendations sent to the prescriber.
‘For pharmacists, managing a gene–drug interaction is fairly similar to managing a drug–drug interaction, and that’s already very common for them. So, pharmacogenetics really nicely connects to the knowledge they already have,’ says Swen.
‘There is testing in community pharmacy in the US, Canada, Cyprus, the Netherlands and Australia, but not yet in the UK,’ says Tim Rendell, head of pharmacy at Day Lewis, a UK pharmacy chain. ‘Currently in the UK, it’s not understood by community pharmacists at all.’
As part of a professional doctorate, Rendell is developing a service specification for a pharmacogenomics testing service in England that he is aiming to trial in pharmacies later this year. ‘This is definitely a service that every community pharmacy could be offering. I think a few years out, when the testing price comes down even further, it will become very much routine,’ he says. He adds that, to begin with, it would be best to focus on offering tests for specific single genes to people taking several medications or starting new treatments.
A report from the Royal College of Physicians and the British Pharmacological Society published in March 2022 offered strategies to overcome the barriers that are holding back implementation, including offering remote support from pharmacogenomics specialists for patient-facing clinicians. Despite the challenges, the report concludes that testing could save the health service money by reducing the £530 million annual cost of hospital admissions for drug side effects.
Even though the idea of personalised prescriptions based on our genes still sounds like something from science fiction, the truth is that it is remarkably simple and straightforward thanks to low-cost DNA sequencing. Yet, despite this and the abundance of evidence in its favour, these tests are still hard to come by. Until this changes, patients will continue to receive the wrong dose of antidepressants, get no relief from a painkiller, or even die from what should be a life-saving treatment.
Ian Le Guillou is a science writer based in London, UK













No comments yet