When it’s a ribozyme. Clare Sansom reports

One of the first principles taught to elementary students of biochemistry is that enzymes – the catalysts that can speed up chemical reactions in living systems millions of times – are proteins. This is certainly true for the vast majority of enzymes, but it is a simplification – just as the traditional statement of the central dogma of molecular biology (DNA makes RNA makes protein) is a simplification. The surprising discovery, in 1982, that RNA molecules alone can have catalytic activity proved for the first time that nucleic acids are more than mere passive carriers of information. We now know that RNA catalysis is essential to life; we understand the chemical mechanism of many of these ‘ribozymes’ and have begun to re-design and engineer them, initially mainly as research tools but with an eye on applications as switches and biosensors and, eventually, as therapeutics.
The discovery of ribozymes showed once and for all that RNA is a central participant in the life of the cell
Thomas Cech of the University of Colorado in the US was the first to discover an RNA molecule with enzymatic activity. In the late 1970s, he had just set up his own lab at Colorado and was studying nucleic acid processing and gene expression in a pond-living protozoan called Tetrahymena thermophila. He noticed that RNA molecules from this little-known organism could be cut into pieces, or spliced, when isolated in a testtube. ‘We wasted a year looking for proteins involved in this splicing reaction. Because it was so fast and specific it had to be catalysed by an enzyme, and of course we knew that “all enzymes are proteins”,’ he says. ‘It took us a long time to realise that there was no protein and it was the RNA itself that was the catalyst.’ He set about to prove this ‘heresy’ by expressing the same RNA molecule in Escherichia coli and showing that no contaminating Tetrahymena protein could be responsible for the reaction.
Cech was also responsible for coining the name ‘ribozyme’ when he organised a competition in his lab to name the molecule they had discovered. ‘Several of the suggestions we had played on the name Tetrahymena. I chose the more general “ribozyme” but I was taking a risk because ours was still the only example known. We would have looked rather silly if it had turned out to be the only example, or perhaps only found in protozoa,’ he explains. Of course, these fears turned out to be groundless: the second catalytic RNA was discovered when Sidney Altman of Yale University in the US showed that the catalytic part of an RNA–protein complex found in bacteria, RNase P, was in fact the nucleic acid.
Cech and Altman shared the 1989 Nobel prize in chemistry for these discoveries, and we now know of ribozymes that are involved in many different biochemical processes: mainly, but not only, in RNA processing. ‘The discovery of ribozymes showed once and for all that RNA is a central participant in the life of the cell, and led to a burgeoning interest in RNA and several more Nobel prizes,’ says Cech.
Take a hammer to it
The so-called ‘hammerhead’ ribozymes – named after a perceived resemblance between early two-dimensional diagrams of their structure and the shark – catalyse site-specific cleavage of RNA molecules. They were first observed in the mid-1980s in tiny, virus-like plant pathogens called viroids, but are now known to occur throughout nature, including in humans. The structure of these fairly small RNAs and the chemistry of the simple reaction they catalyse is now very well understood. Their natural function is not, strictly speaking, ‘catalysis’ at all: the target RNA sequence is cleaved by the isomerisation of the phosphodiester bond, with the ribozyme itself being consumed by the reaction. It was originally assumed that this reaction required a divalent metal ion, much as a metalloprotease enzyme requires zinc, but these ions are now thought to be principally involved in stabilising the RNA scaffolds.
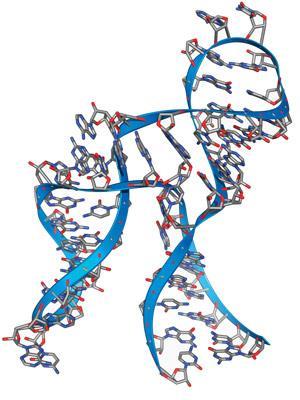
Hammerhead ribozymes are intrinsically cis-acting: that is, their natural activity is to cleave and hence to inactivate themselves. As they are quite small, simple molecules, it is relatively easy to engineer them to cleave external substrates (trans-acting). Furthermore, a hammerhead ribozyme consists of a conserved, central catalytic core with two ‘arms’ that recognise the RNA sequence to be cleaved, so it is fairly straightforward to design a ribozyme to cleave a specific sequence by altering the nucleotides in the arms.
Several groups are using this technique to design ribozymes to recognise and cleave specific RNA sequences, both to help elucidate molecular mechanisms in vivo and for biotechnological applications. Matasora Fukuda and his group at Fukuoka University in Japan are using it to understand some of the mechanisms involved in RNA editing. This name is given to processes that change the chemistry of the bases in RNA during or after their transcription from DNA. Adenosine-to-inosine (A-to-I) RNA editing, as the name implies, involves the deamination of adenine to form another purine, hypoxanthine (inosine is the hypoxanthine nucleoside). Since adenosine is decoded as guanine, it can alter the protein sequence when it occurs in protein-coding regions, and it regulates a variety of biological processes.
Fukuda and his co-workers used the generic fact that hammerhead ribozymes can cleave the base sequence UAA but not UGA (U: uracil; A: adenine; G: guanine) and the similarity between guanosine and inosine to engineer a ribozyme structure that cleaves an RNA editing target site only if the A-to-I substitution has not been made. They then created a library of ribozymes containing this central motif extended on each side with random sequences, and selected the most stable and therefore most active one for further testing. ‘This ribozyme cleaved unedited RNA sequences but not edited ones, but it was fully active only at higher magnesium concentrations than those that occur in cells,’ says Fukuda. ‘Creating a ribozyme that works at physiological salt concentrations will require further modification.’ Aberrations in A-to-I RNA editing have been observed in several diseases, and Fukuda hopes that novel nucleic acid-targeted therapies may one day be created using similar techniques. However, it is not yet clear whether these aberrations are a cause or an effect of disease. ‘The first applications of these techniques are likely to be novel tools for molecular biology research,’ adds Fukuda.
The first applications are likely to be tools for molecular biology
Robert Penchovsky of Sofia University in Bulgaria is using a similar approach to design ribozymes that detect and respond to the presence of specific small molecules. An aptamer is a short nucleic acid sequence or a small protein that binds to a given molecular target. Most aptamers are synthetic, and allosteric ribozymes – ones that are active only with a ligand bound outside the catalytic domain – can be designed by fusing an RNA aptamer containing the binding site with, for example, a catalytic hammerhead ribozyme.
Penchovsky used computer modelling to design molecules of this type in which the ribozyme changes conformation between active and inactive states in response to theophylline, a purine that is chemically similar to caffeine. ‘These simple ribozymes can act as molecular logic gates with the Boolean logic YES function if theophylline binding activates the ribozyme and the NOT function if binding deactivates it,’ says Penchovsky. ‘I can also design oligonucleotide-sensing ribozymes that act as AND and as OR gates, and similar biosensor systems have already been used in high throughput screening arrays for antibacterial drug discovery.’ There are other potential applications of this technology in molecular computing and antibiotic design, and Penchovsky is seeking industrial collaborators to develop these further.
The ribosome
Hammerhead ribozymes are fairly small, tractable molecules that can be synthesised relatively easily. There are other biologically important ribozymes, however, that are right at the other end of the macromolecular spectrum. The ribosome, the ‘molecular machine’ that catalyses protein synthesis and is found in all living cells, has two subunits that only come together during protein synthesis, and each subunit includes many protein and RNA molecules. When the first high resolution structure of the large subunit was published, the researchers involved, Tom Steitz and his group at Yale, commented that there were proteins everywhere on the subunit’s surface ‘except in the active site where peptide bond formation occurs and where it contacts the small subunit’. Steitz and his co-workers concluded that, very unexpectedly, it was the RNA molecules that were principally, if not solely, involved in peptide synthesis, with the proteins acting as ‘staples’ to hold them in place. They had established that the ribosome is a ribozyme.
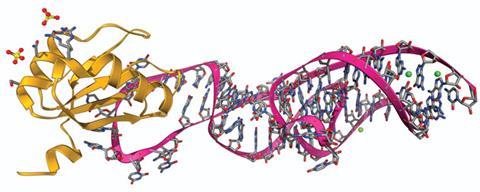
Venki Ramakrishnan from the MRC Laboratory of Molecular Biology in Cambridge, UK, shared the 2009 Nobel prize in chemistry for studies of the ribosome with Steitz and with Ada Yonath of the Weizmann Institute in Israel. He has devoted much recent research to understanding the mechanism through which the ribosome recognises the three nucleotides that form a ‘stop codon’, terminates protein synthesis and releases the newly formed protein. The standard genetic code uses three stop codons, UAA, UGA and UAG, and Ramakrishnan’s remarkable structural studies – which involved electron microscopy as much as crystallography – have shown how a eukaryotic protein known as release factor can, when bound to the ribosome, use base stacking and hydrogen bonding to recognise these base triplets but no others. It is interesting that this, too, can be compared to a Boolean logic gate, in this case a NAND gate with G represented by 1 and A by 0. Any combination of these two nucleotides except ‘GG’ (‘11’) in the second and third codon positions will signal the termination of protein synthesis.
These discoveries, and others, show that RNA molecules are able to catalyse the complete minimal set of chemical reactions that is essential for life, and this has led to a theory on the origin of life termed the ‘RNA world’. This states that, since RNA molecules are able to self-replicate and to combine a DNA-like function of information storage with a protein- like function of chemical catalysis, all life on earth could have evolved from primitive ‘organisms’ comprised of RNA only. ‘It is impossible to prove this theory since there are no molecular fossils, but all the information we have accumulated about ribozymes and RNA suggests that it is not only possible but likely,’ says Cech.
Gaining control
It is not even possible to imagine scientists creating a ribozyme that is structurally and functionally as complex as the ribosome any time soon. But researchers are already tinkering with natural ribosomes to subtly alter their properties. Initially, this was limited by their inability to control which out of a pool of large and small ribosomal subunits available would form active complexes. Now, however, several groups, including Jason Chin’s at the MRC Laboratory of Molecular Biology, Alexander Mankin’s at the University of Illinois and Michael Jewett’s at Northwestern University in the US have shown that it is possible to design fully orthogonal ribosomes with subunits that have been joined by covalent linkers and that are still catalytically active.
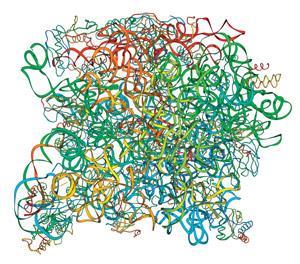
Mankin, Jewett and their teams have designed a ‘tethered’ ribosome in which one RNA component of each of the large and small subunits of the E. coli ribosome were joined by two poly-adenosine nucleotides. ‘These linkers had to be long enough to ensure that the subunits could make the whole range of conformational changes necessary for protein synthesis, but short enough to prevent either subunit from associating with another ribosome,’ explains Mankin. ‘One eight-nucleotide and one nine-nucleotide linker work very well.’ These ribosomes, termed Ribo-T, can catalyse protein synthesis in E. coli cells even if there are no wild-type ribosomes present, although bacteria with no wild-type ribosomes grow only half as fast. ‘We are using Ribo-T to explore the mechanism of protein synthesis even further, and we will be able to engineer tethered ribosomes with unusual functions, eventually even coaxing them to synthesise polymers other than proteins,’ adds Jewett.
The ribosome is not the only large molecular machine that has been shown to be a ribozyme. The genes of most eukaryotes contain non-coding segments called introns that must be removed from the transcribed messenger RNA before that can reach a ribosome. This process, in which the RNA is cleaved at the start and end of each intron and then the protein-coding exon sequences are re-joined, is termed splicing and the protein–RNA complex that catalyses this reaction is the spliceosome. It consists of five small RNA molecules, protein complexes and magnesium ions and again, the catalytic unit is formed from RNA and stabilised by the proteins and ions. The structure of the RNA component of the spliceosome is very similar to that of a much smaller ribozyme, the self-splicing group II intron that is found in the genomes of all organisms. ‘Proteins stabilise the structure of group II introns and enhance their activity, although they are not necessary for catalysis,’ explains Kiyoshi Nagai of the MRC Laboratory of Molecular Biology. ‘Recent structural studies of the spliceosome have revealed even more similarities between the spliceosome and group II self-splicing introns, indicating a possible common evolutionary origin.’
Tinkering with the structure and function of large and small ribozymes has already generated many new insights into molecular biology and molecular structures with novel chemistry and functions. They are certain to play an increasingly important part in the emerging discipline of synthetic biology.
Clare Sansom is a science writer based in London, UK










No comments yet