A new computational technique allows researchers to see clearer images of biomolecules by breaking the atomic force microscope’s (AFM) usual resolution limits. The method offers a new way to obtain high quality structural data on biological compounds like proteins in their native environments.
While x-ray crystallography and cryo-electron microscopy allow researchers to image biomolecules at the atomic level, these techniques rely on samples that have been crystalised or frozen at ultra-cold temperatures, which can alter the molecules’ natural shapes. AFM allows molecules to be probed under more normal physiological conditions, but the images are not as sharp as with the other methods.
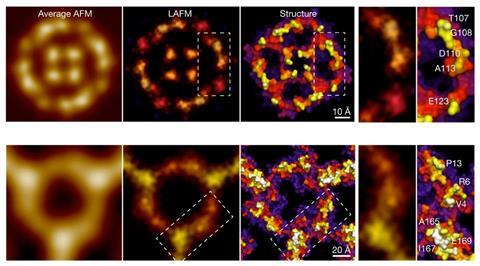
Now, a team led by Simon Schuering from Cornell University, US, has found a way to improve the quality of AFM images. The strategy, which was inspired by methods already used in optical microscopy, uses a localisation image reconstruction algorithm to process data from multiple scans of single molecules. It can even be used retroactively to reveal new details hidden in old AFM data.
Demonstrating the technique, the researchers explain that they could ‘increase the resolution beyond the limits set by the tip radius, and resolve single amino acid residues on soft protein surfaces in native and dynamic conditions’.
Emphasising another benefit of the technique, Scheuring told Weill Cornell Medicince’s website that using highly resolved AFM to monitor single molecules as they move between confirmations could help to ‘avoid the potentially misleading results that can occur when averaging data from many molecules’.













No comments yet