Editor’s note: The Nature Chemical Biology paper that this news story is based on has been retracted.
A combined computational and biochemical study has led to the discovery of a new type of riboswitch – RNA elements that sense cellular metabolites – which can directly and selectively bind iron(II) ions and regulate protein production accordingly.1 The findings offer new possibilities for the design of RNA-based biosensors.
Despite iron’s biological importance, knowledge of how cells can detect this metal has been limited to protein-dependent mechanisms. ‘Iron-binding proteins have been the best-studied iron sensors so far,’ notes Arati Ramesh from the National Centre for Biological Sciences in India who led the study. ‘With the discovery of Sensei (Sense iron) RNAs, the responsibility of iron sensing and modulation is now extended to RNAs.’
Iron plays a central role in processes such as cellular respiration or DNA damage, so knowing how cells sense this metal is crucial in biochemistry. In the last few years, scientists have shown that RNAs can act as sensors for a range of metals. Several metal-ion-binding RNA families, which specifically sense magnesium, manganese and nickel/cobalt (NiCo) ions, have been discovered. Now, a new family can be added to the list, suggesting that metalloregulation by RNAs is more widespread than previously thought.
‘The authors have discovered that riboswitches may directly sense iron and regulate gene expression without the apparent help of protein factors,’ comments biochemist Daniel Lafontaine at the University of Sherbrooke in Canada. ‘This is important as it expands the previously known repertoire of cellular metabolites bound by regulatory riboswitches.’
At the beginning of the study, the team looked at NiCo riboswitches and found variants that had mutations in nucleotides that are key to cobalt binding. ‘These variants were found near genes encoding for iron-related proteins, raising a possibility that they could bind iron,’ Ramesh says. ‘We took them as a starting point and used the base-pairing information and nucleotide frequencies to carry out a search in bacterial genomes.’ This bioinformatic analysis allowed the researchers to identify the new Sensei RNAs.
They then synthesised six representatives of this family and studied their interaction with iron. ‘To test our findings, we kept the RNA and iron in two chambers separated by a membrane that allows only iron to pass through,’ explains Ramesh. When the iron passed through and bound the RNAs, binding was detected through a colour change indicating iron’s strong affinity for the riboswitches. Ramesh adds that Sensei RNAs are not restricted to any one group of organisms.
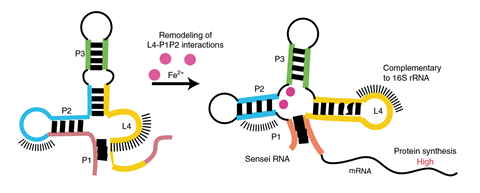
‘Iron-sensing riboswitches are relatively simple structured RNA molecules that exhibit a four-way junction or a cloverleaf structure,’ explains Lafontaine. ‘Upon the binding of iron, these RNA molecules are reorganised into a structure allowing protein synthesis of the downstream gene.’
The team then engineered the metal selectivity of RNAs, converting a NiCo riboswitch into an iron-sensing molecule and making a Sensei RNA detect cobalt instead of iron. ‘This opens avenues to the design of Sensei-based biosensors.’ Ramesh says. The researchers now want to understand the mechanism by which the new switches increase protein translation.










1 Reader's comment