Understanding protein folding and stability leads to new proteins with hopes of creating structures that can perform novel chemistry

Scientists in the UK and US have designed and synthesised unnatural protein structures, using theoretical calculations to explore the factors affecting protein folding and stability. They hope their approach can ultimately be used to create proteins with new biological or chemical applications.
The shape into which a protein folds is crucial to its function, with several severe diseases such as Alzheimer’s thought to be the result of misfolded proteins. However, while the chemical forces that govern protein folding are understood, the intricacy with which they compete and interact means that predicting how a particular protein will fold from its amino acid sequence is extremely difficult. Now, two separate research groups have derived rules governing the stability of a particular class of proteins and used these rules to predict and synthesise stable structures not found in nature.
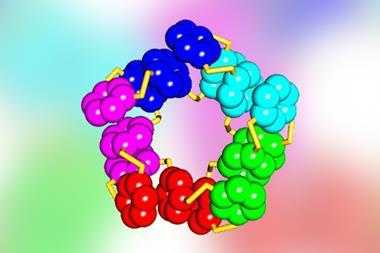
Both groups looked at a-helical coiled coils, which are made up of coiled chains of amino acids, themselves coiled around one another into a ‘supercoil’. The structure is characterised by relatively few parameters, such as the twists on the coils and their dimensions. Therefore the structures of proteins with this shape are relatively well understood, and several new proteins have been designed, although these have mainly mimicked natural structures. The two groups – the first led by David Baker at the University of Washington in Seattle, US, the second by Derek Woolfson at the University of Bristol, UK – set out to unlock what makes a particular structure stable or unstable.
Searching for stability
Baker’s group sought repeating coiled coil structures in which, after a fixed number of twists, both sets of coils arrive back where they started. Structures where one set of coils has to be stretched or compressed from its natural curvature to achieve this store potential energy like springs, and are therefore less stable, so Baker’s group looked for structures that were as unstrained as possible. Even if a structure is stable, whether or not a protein adopts it depends on its amino acid sequence, as various factors come into play such as hydrophobic interactions. The researchers used a computer algorithm to search for amino acid sequences that would adopt these unstrained structures. When they found suitable sequences, they synthesised them experimentally. Interestingly, they found that many of their synthetic proteins were more stable to heat and chemical denaturation than natural ones.1
Woolfson’s group focused on a variant of coiled coil structures called a-helical barrels, in which five or more helices are bundled in cylinders around a central channel. They found amino acid combinations that stabilise helix–helix interactions in simpler and more common natural proteins with just two helices. Then they grafted these onto computational models of entirely new a-helical barrels to create the multiple helix–helix interactions needed, thereby designing theoretically stable versions of the structures. They chemically synthesised 22 of these proteins.2
Both groups found many structures not seen in nature, and they are interested in whether they can can do things that are beyond the capabilities of natural proteins. Woolfson’s group, for example, hopes to use the holes in their structures to produce ion channels or to engineer pores into cells. ‘Proteins carry out essentially all the important processes in the body,’ says Baker. ‘The goal of protein design is to figure out new proteins that solve today’s problems as well as naturally-occurring proteins solve the problems that came up during evolution.’
William DeGrado of the University of California, San Francisco in the US says the systematic way in which both groups have worked out many of the rules governing stability has thrown up some intriguing findings. He suggests, for example, that Baker’s finding that natural proteins are rarely the most stable possible structures suggests that they have evolved towards ‘intermediate stability’, perhaps so they can be taken apart in the human body.










No comments yet