Researchers have developed new ways of simulating the folding of membrane proteins, which could aid drug development
Membrane proteins are important targets for drug development, yet notoriously difficult to study by both experimental and theoretical methods. Researchers in Italy and the UK have now developed a computational approach that can simulate the folding of membrane proteins in atomic detail.1
Membrane proteins need a membrane to attain their native folded structure. This means they are even harder to crystallize than other proteins for crystallographic purposes. It was also thought that, for computational analysis, the movements of lipid molecules of the membrane would have to be computed as well.
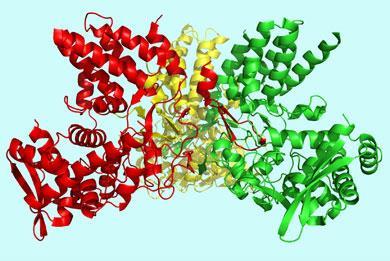
Jakob Ulmschneider at the University of Rome, together with his brother Martin at the University of Oxford and other colleagues think that the best approach is to replace the membrane by a very simple sheet designed to be very hydrophobic in the middle and more hydrophilic at the surfaces. They express this mathematically as a gradient running across the thickness of the membrane. On the protein side, so-called Born radii have to be calculated in order to reflect the interaction between the protein atoms and the surrounding medium.
Monte Carlo or bust
With this simplifying assumption known as an ’implicit membrane’, the researchers can afford to spend most of their computing power on the actual protein of interest and study its movements at atomic resolution. ’Similar implicit membranes have been developed by Wonpil Im and Charles Brooks at the Scripps Research Institute in the US to be combined with molecular dynamics simulations,’ said Martin Ulmschneider. ’In contrast, our study uses so-called Monte Carlo simulations, which apply millions of random steps to optimize structures for any given environment.’
Having previously tested their method on small model peptides,2 the Ulmschneider brothers are now applying it to large parts of real proteins. Specifically, they studied the transmembrane protein domains of the influenza A virus, and the sarcolipin protein from human muscle. For these proteins, whose structures are already known, they could show that the simulations produce biologically meaningful results that are in agreement with experimental findings.
Ernst-Walter Knapp from the Free University at Berlin, Germany, praised the work: ’They combined an efficient Monte Carlo approach with implicit models for membrane and solvent, overcoming the problem of an exaggerated structure stability by proper parametrization,’ Knapp told Chemistry World. ’This is a most efficient simulation scheme for membrane proteins, opening avenues to explore folding and stability of large membrane proteins soon.’
Alrik Koppenh?fer, a partner at the Life Science Consulting Networkbased at Heidelberg, Germany, also welcomed the development. ’Computing simulation of the folding of membrane proteins could seriously help scientists develop new and better drugs for diseases like Alzheimer or diabetes. The Ulmschneiders’ findings are very encouraging and should be much welcomed by the industry,’ he told Chemistry World.
Michael Gross
Enjoy this story? Spread the word using the ’tools’ menu on the left.
References
1 J P Ulmschneider, M B Ulmschneider and A Di Nola, Proteins, DOI: 10.1002/prot.21519
2 M B Ulmschneider et al, Biophys. J., 2007, 92, 2338




No comments yet