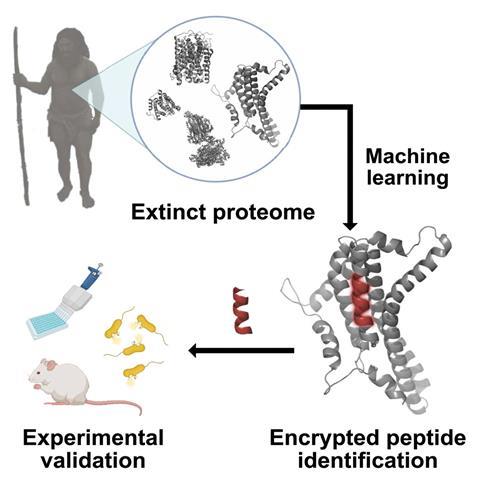
A combination of computational and experimental techniques unveils ‘encrypted’ peptides in proteomes of ancient humans with potential as antibiotic agents
Neanderthal proteins could be a rich new source of medicinal molecules. The finding comes from researchers who have pioneered a technique called ‘molecular de-extinction’ by analysing the proteomes of ancient hominins. The work has already uncovered a series of peptides with antibiotic activity.
‘Molecules from the past [could] perhaps provide solutions to present-day problems,’ says César de la Fuente, from the University of Pennsylvania, US, who led the project. ‘It’s like Jurassic Park, but we just revive small molecules,’ he adds. But unlike attempts to bring back whole extinct organisms like those in Michael Crichton’s fictional dinosaur theme park, de la Fuente and his co-workers note that their strategy of reviving individual biomolecules brings fewer ethical problems and could bring important technological advantages.
De la Fuente’s team had previously developed an algorithm to mine the human proteome to look for ‘encrypted’ peptides – fragments that detach from bigger proteins. ‘Although we still ignore the basic biological functions of encrypted peptides, it’s possible they play a key role in the immune system,’ he says. ‘We had also discovered some peptides with antibiotic properties in blood plasma proteins and other parts of the [modern] human proteome.’
Encouraged by these findings, de la Fuente’s team decided to delve into the proteome of humankind’s closest relatives: the Neanderthals and Denisovans. Despite the scarcity of sequence sources – data availability relies on well-preserved samples of mitochondrial DNA – the researchers were able to identify 69 promising peptides, of which six showed antibiotic activity against different pathogens in mouse models.
Jean-Louis Reymond, a drug discovery expert at the University of Bern, Switzerland, notes that the potential antibiotics aren’t particularly potent. However, he describes the work as ‘excellent biology’ and says that the evidence of in vivo activity and moderate toxicity is ‘in principle a promising outcome’.
Another interesting aspect is that the active peptides are quite different in sequence from known antimicrobial peptides, says Reymond. The ancient peptides have higher amount of polar, acidic, and aromatic amino acids, and lower levels of basic residues.
‘[These] unusual amino acid distributions could reflect differences in the selective [evolutionary] pressures that acted on these proteins in the past,’ says Laureano Carpio, a researcher at drug discovery company ProtoQSAR, in Valencia, Spain.
However, Carpio notes that this observation could be skewed by the small sample size. ‘It’s also possible that the peptides identified in this study don’t represent the full range of peptides that existed in extinct organisms,’ he says. ‘In the future, identifying additional peptides could allow us to study the evolution of these molecules over time.’
According to De la Fuente, the de-extinct peptides appear to preferentially target the inner membrane of Gram-negative bacteria. This is in contrast to modern encrypted peptides that tend to target the outer membrane, a structure that blocks many antibiotics, including penicillin. De la Fuente describes this observation as ‘almost anecdotal’, but he believes further research could confirm this theory, which could help the design of drugs against traditionally resistant microorganisms. ‘The results are really promising,’ he adds. ‘One of the de-extinct peptides performs particularly well against skin infections in mice, comparable with state-of-the-art solutions like polymyxin.’
Considering the current crisis of antibiotic resistance, finding effective new drugs is a hugely important area of research. ‘Certainly, the de-extinction of peptides and other biomolecules presents an intriguing avenue for discovering novel antibiotics,’ says Carpio. ‘This study marks a pivotal starting point for such exploration.’
References
J R M A Maasch et al. Cell Host Microbe, 2023, 31, 8, 1260 (DOI: 10.1016/j.chom.2023.07.001).
















1 Reader's comment