Microfluidic device allows speedy susceptibility testing so the right antibiotic can be given
A microfluidic chip that can potentially find out whether bacteria are susceptible to an antibiotic within 30 minutes has been developed by researchers in Sweden, who say it may help in the fight against antimicrobial resistance by helping doctors to choose the correct antibiotic.
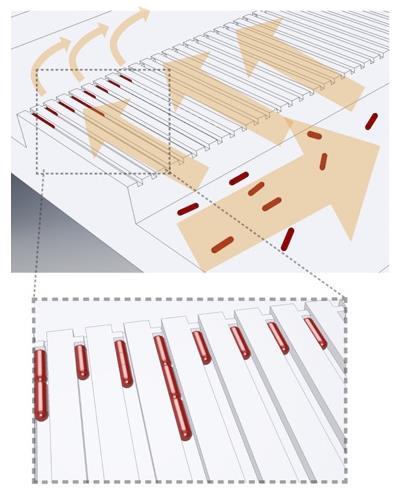
Current antibiotic susceptibility tests are far too slow for doctors to delay treatment until the results are known. The current susceptibility test for a urinary tract infection – for example – involves culturing the urine on agar plates in the presence of different antibiotics to see whether bacterial colonies form. Doctors must therefore often choose a narrow-spectrum antibiotic that may be ineffective or a broad-spectrum one that may have more severe side-effects, and may contribute unnecessarily to resistance.
Urinary tract infections affect around 100 million women every year and show widespread antibiotic resistance. However, ‘It takes approximately two days before you know whether it’s a resistant infection,’ says biophysicist Johan Elf of Uppsala University. He and his colleagues designed a microfluidic device that aims to speed up the process.
Their plastic device has two rows, each comprising 2000 micrometre-scale tubes with a constriction at one end. When infected urine flows through the device, bacteria are captured in the tubes. The researchers then flush growth medium through the device, with the medium in one row containing an antibiotic.
Even the fastest growing bacteria take around 30 minutes to divide but, using image analysis techniques it has developed for its basic research, Elf’s team monitored the volume extension of individual cells as they grew. By comparing the average heights of the bacteria in the tubes washed with the antibiotic-infused and antibiotic-free growth media, the researchers could determine after 3-11 minutes’ growth whether they were susceptible to various antibiotics. The total analysis time, including sample loading and readout, was less than 30 minutes. The researchers blind tested 49 urine samples infected with E. coli from Uppsala University Hospital pathology lab: they correctly classified every sample as either susceptible or resistant to the antibiotic ciprofloxacin. They believe the technology could be expanded to analyse other kinds of infectious bacteria, such as cerebrospinal fluid in meningitis patients. A commercial company is now working to make a mass-producible, user-friendly version suitable for primary care. ‘It’s challenging, because there are no commercial microfluidic devices that I know of containing such small structures,’ says Elf, ‘But I’m pretty confident this can be mass produced.’
Chemical engineer John Oakey of the University of Wyoming in the US describes the device as ‘a great example of how microfluidics can be combined with emerging assays and single cell tools to create opportunities in precision or individualised medicine’. Clinical microbiologist Omai Garner of University of California, Los Angeles cautions that sample contamination issues would make testing in primary care difficult: ‘The pre-analytic processing of samples before they’re tested is extensive within a laboratory setting but non-existent at the point of care,’ he says. Nevertheless, he says: ‘A device or something like this should be available within the next five to ten years.’
References
O Baltekin et al, Proc. Natl. Acad. Sci. USA, 2017, DOI: 10.1073/pnas.1708558114











No comments yet