The transcription factor FOXP3’s interactions with DNA present more evidence of the importance of disorder
Since its earliest days, supramolecular chemistry has taken inspiration from biology. To create a ‘chemistry beyond the molecule’, supramolecular chemists can learn from the way nature builds hierarchies of organisation from the selective and orderly interactions of molecular components. At least, that’s what Jean-Marie Lehn and I argued in an overview of the subject in 2000.1 Yet while I still believe that today, I’m less sure that nature’s molecular principles can be easily translated into what Lehn has called a rational ‘science of informed matter’2 – and even less so that the principles used in supramolecular chemistry to create wonderful edifices of molecular order and design will by themselves give us anything like proto-living systems.
The reason is that life’s molecular principles are far less transparent than we thought even a few decades ago, and certainly less amenable to rational bottom-up design. An example is supplied by a new study of how a transcription-factor protein called FOXP3 interacts with DNA to influence the differentiation of regulatory T (Treg) cells, key components of the immune system, from their precursor cells. Transcription factors regulate gene expression, and one way FOXP3 seems to do this is by binding directly to DNA as dimers in which two of the proteins sit in ‘head-to-head’ contact.
But Wenxiang Zhang, Fangwei Leng and their colleagues at Boston Children’s Hospital reported in November that FOXP3 can stick to DNA in a very different and decidedly weird way.3 They showed that the protein can bind two double helices together via five pairs of the proteins acting as the rungs of a ladder-like complex bridging the two strands: a complicated supramolecular structure for which I know of no biological precedent, almost mimicking at larger scale the ladder-like unions of base pairs holding each double helix together.
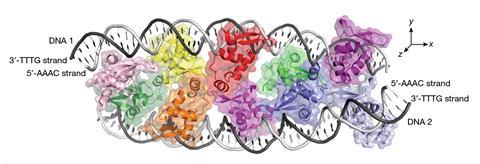
It’s weird that FOXP3 has this alternative, biologically relevant mode of action. It’s doubly weird that the five dimer units stick to sections of DNA previously thought to be meaningless junk: ‘microsatellites’ in which short sequences of a G base and a few T’s (TnG, such as T3G) repeat several times. TnG repeats ‘are usually considered non-functional and are discarded by sequence-analysis programs’, say immunologists Zhi Liu and Ye Zheng in a Nature commentary,4 which prompts me (not least because this isn’t an isolated example) to propose a new year’s resolution for molecular biologists: stop assuming we know which parts of DNA matter and which don’t.
The ladder complex is vital to the proper functioning of Treg cells: if the FOXP3 gene is mutated to disrupt rung formation, the cells aren’t so good at keeping the immune system in line. It’s a fair (but unproven) assumption that this tethering of two pieces of DNA affects the three-dimensional organisation of chromatin (the fibrous material of the chromosomes), perhaps securing an ‘enhancer’ sequence close to the gene it regulates and thereby altering gene expression. Yet those roles evidently don’t depend on precise programming: FOXP3 will stick to a range of TnG microsatellites and can adopt various inter-rung spacings.
So far, so perplexing. But get this: other studies have shown that FOXP3 will form huge multiprotein complexes with as many as 361 other proteins in Treg cells, including some that regulate the expression of FOXP3 itself and others whose expression FOXP3 regulates.5 It seems almost as if everything affects everything else in a vast network, and that how FOXP3 interacts with DNA is just one part of a much bigger picture.
What hope do we have of making sense of all this? It’s an understatement to say it’s complicated. But I don’t despair. Quite the opposite: I believe the FOXP3 story reinforces several considerations we have already learnt about life’s molecular mechanisms.
First, those often do not rely on the kind of specificity of structure and interaction typical in supramolecular chemistry – especially insofar as they pertain to gene regulation and the assignment of cell fates. Rather, the rules are fuzzy and combinatorial, such that different collections of molecules can induce the same outcomes, and the same collections can induce different outcomes depending on context. Cognition, not computation or clockwork, is a better metaphor for how the cell’s decisions are made.
And the molecules are ‘designed’ by evolution to work in just this way. It might be puzzling, for example, if a precisely structured FOXP3 protein could do such different things with such wide tolerances – but it is surely because the protein is not so structured, but (like many transcription factors) has disordered regions, that it can show such versatility. There is logic in life, but it is fuzzy, analogue, and barely understood – and we’re far from being ready to design molecular systems along the same lines.
References
1 J-M Lehn and P Ball, in The New Chemistry (ed. N Hall). Cambridge University Press, 2000
2 J-M Lehn, Supramolecular Chemistry. Wiley-VCH, Weinheim, 1995
3 W Zhang et al, Nature, 2023, 624, 433 (DOI: 10.1038/s41586-023-06793-z)
4 Z Liu and Y Zheng, Nature, 2023, 624, 255 (DOI: 10.1038/d41586-023-03628-9)
5 D Rudra et al, Nat. Immunol., 2012, 13, 1010 (DOI: 10.1038/ni.2402)





![[Br4F21]- index image](https://d2cbg94ubxgsnp.cloudfront.net/Pictures/380x253/1/7/3/532173_br4f21indeximage_49094.jpg)
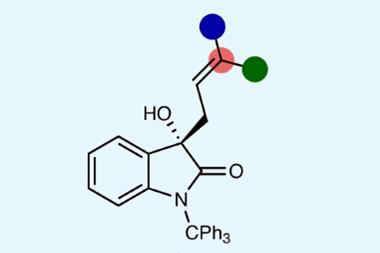


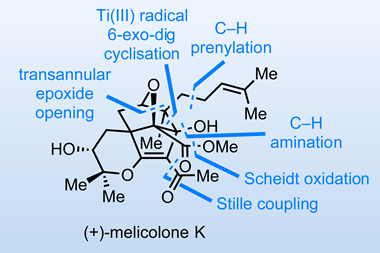






1 Reader's comment