Scientists in the US have devised an enzymatic logic system that could be used for releasing drugs.1 This is the first report of a man-made biomolecular system that can process a series of physiological signals, without the use of electronics.
Biocomputer-based logic systems that process biomolecular signals could revolutionise drug administration. By harnessing signal-responsive electrode surfaces that respond to biochemical signals, widespread personalised medicine takes a step closer to reality.
Historically, drug-release systems have been plagued by slow and uncontrolled release. Various external triggers, including temperature, pH and biochemical species, have been used to stimulate drug release. Systems activated by biochemical signals are often complicated and limited, combining both the receptor and the release system. Physical separation of these two components on individual electrodes would simplify the process.
Expanding on their recent work with glucose sensing electrodes,2 Evgeny Katz and Shay Mailloux at Clarkson University, in collaboration with Jan Halámek from the State University of New York at Albany, have developed a logical biomolecule release system. An electrode covered by a redox-active, iron(III)-cross-linked alginate polymer film with physically entrapped biomolecules serves as the substance-releasing component and a pyrroloquinoline quinone (PQQ)-modified electrode serves as the biocatalytic electrode.
The system is highly selective as it responds only to specific combinations of biomolecular input signals that are processed by carefully designed combination of logic gates. Upon oxidation of nicotinamide adenine dinucleotide (NADH), the biocatalytic electrode produces a negative potential and reductive current. In turn, the iron(III)-cross-linked ions are reduced to iron(II), the film becomes soluble and the entrapped biomolecules are released. Using NADH to trigger the release is important as it can be generated by numerous biocatalytic systems, imparting wide applicability to the model system.
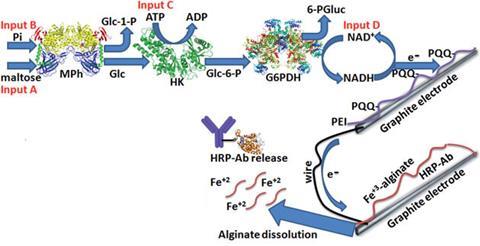
Katz says the novelty of their approach is the release of the biomolecule on receiving particular signals, which could eventually be those from a patient’s body. He emphasises that this is the first system integrating biocomputing with biomolecule release: ‘The study demonstrates the use of biocomputing – in other words, information processing by biochemical means – for triggering another process. This might be compared to connecting a computer with a printer. In this combination the computer is processing the information and the printer is printing the results. Our biochemical system conceptually works in this way.’
A Prasanna de Silva, a molecular computation expert at Queens University Belfast, UK, describes the system as a rare example of logical drug release. While, professor in unconventional computing at the University of the West of England, UK, Andy Adamatzky says that ‘Katz's lab has opened new application domains for future and emergent computing paradigms. They transform unconventional computing from a purely theoretical field to a vibrant experimental area where concepts of unorthodox information processing and decision making can be realised with biomolecules and, potentially, applied to personal illness treatment.’
Although this elegant concept is not ready for real medical applications just yet, work is already underway using real biofluids in pursuit of this goal.





No comments yet