Healthcare tailored to suit the genetic makeup of the patient is finally coming to fruition, as Anna Lewcock reports
Healthcare tailored to suit the genetic makeup of the patient is finally coming to fruition, as Anna Lewcock reports
Ten years ago, two men stood with the US president in the White House and announced the culmination of a colossal project involving the combined power of over 1000 scientists and years of research - the findings, they said, marked a new starting point for science. The elder of the two, genetics luminary Craig Venter, stepped forward and before the assembled media declared: ’today, 26 June, in the year 2000, marks an historic point in the 100,000-year record of humanity. We’re announcing today, for the first time our species can read the chemical letters of its genetic code.’

The completion of the first draft of the human genome was heralded as the biggest scientific breakthrough of the 21st century, and was the result of 10 years’ work on the Human Genome Project - and $2.7 billion (?1.9 billion) in cold, hard cash. The then UK prime minister Tony Blair hailed the work as ’a revolution in medical science whose implications far surpass even the discovery of antibiotics. A breakthrough that opens the way for massive advances in the treatment of cancer and hereditary disease - and that is only the beginning.’

But a decade later, has the promise and frenzied expectation of the new genomic era come to pass?
The genetics juggernaut
The final version of the human genome, the three billion letters of DNA code that make up the human ’book of life’ was finally completed in 2003. The hope was that by revealing humanity’s genetic blueprint, we’d be able to pin down genetic differences that make individuals more susceptible to certain diseases, or affect the way we respond to therapeutic drugs. It was this second aspect that seemed to promise a future of ’personalised’ medicine, with patients prescribed drugs tailored to their specific genetic profile. Gone would be the days of switching from one drug to another to find a treatment that worked; farewell to unpleasant side effects, tweaking dosages and medical guesswork.
We’re not yet at the stage where a doctor can take a cheek swab or blood test, run a quick assay and be presented with a genetic checklist that dictates the best treatment for your ailment. But research into pharmacogenomics has made enormous progress in the last decade, and continues to move at a staggering pace.

Panos Deloukas leads a group looking at the genetics of complex traits in humans at the Wellcome Trust Sanger Institute in Cambridge, UK, and coordinated the sequencing and analysis of chromosomes 10 and 20 as part of the Human Genome Project. He says technological advances have played a key role in building on the foundation of knowledge laid by the original project. ’If you look back to 2003, trying to genotype 1000 positions was viewed as a very big and costly experiment. Now we can scan a million positions for a fraction of the cost and have the results in a few days.’
Deloukas set up the Sanger Institute’s high-throughput screening facility, and now focuses on identifying genetic variations that seem to affect individuals’ response to drugs. His work is based on genome-wide association studies, which essentially involve rapidly scanning entire genomes and looking out for variations that seem to be linked to the particular response or trait being investigated. Deloukas has spent a lot of time investigating the blood-thinning drug warfarin, one of the most widely used anticoagulants in the world. The drug has what is known as a narrow therapeutic range - the therapeutic dose is very close to a dose that could be dangerously toxic or even lethal - and dose requirements vary significantly from patient to patient. Deloukas has been trying to pin down genetic determinants of this variation and has so far managed to identify three single nucleotide polymorphisms (SNPs - changes in a single nucleotide within a person’s gene sequence) that contribute to warfarin response. ’So what we have been doing is taking these genetic variants together with a number of non-genetic factors like body weight and other medication patients are taking, and developing an algorithm that predicts their optimal dose to initiate warfarin treatment,’ says Deloukas. ’So far this model of genetic and non-genetic factors explains about 60 per cent of the variation that you see in patients’ dose requirements.’
While this is a significant step in explaining the differences in response, and the US Food and Drug Administration (FDA) now recommends lower warfarin doses for some patients based on two of the genetic flags that have been identified, it still doesn’t provide quite enough information to confidently inform patient dosage. Deloukas is hoping that more advanced screening methods will help identify the remaining genetic factors that explain the variation in response to the drug, and help make dose decisions more reliable and safer for patients.
Aside from warfarin, Deloukas also investigates genetic markers that could explain the unpleasant side effects some patients experience when taking certain drugs. He’s been looking at a range of treatments from cholesterol drugs and HIV treatments to a class of blood pressure medicines that seem to trigger persistent coughing in some patients. Genetics, it seems, plays a role in all cases.
Focus on cancer
Beyond Deloukas’ lab, oncology has emerged as a hub of pharmacogenomic research. Several cancer drugs have been developed over recent years that have been found - or designed - to work only in a subset of the patient population who have a particular gene variant.
’Cancers differ from their host - that’s why they’re cancers,’ explains Ian Cree, consultant pathologist and director of the cancer laboratory and the translational oncology research centre at Queen Alexandra Hospital in Portsmouth, UK. ’We have knowledge of pathways that cancer cells use that normal cells don’t, and we are able to hit those pathways with fairly specific agents. If you then find that a particular cancer is dependent on that pathway, you can get some really rather exciting clinical results.’
US pharmaceutical company Amgen’s Vectibix (panitumumab) is an antibody treatment that targets the epidermal growth factor receptor (EGFR), which sits on the cell surface and is particularly important in promoting tumour growth in colorectal cancer. Vectibix stops the growth factors binding to the receptor and stimulating tumour growth.
But while Vectibix was in Phase III studies, research started to suggest that a certain biomarker - a biochemical feature such as a protein or genetic signature - could affect patient response to the drug. It all hung on the KRAS gene, which provides instructions for making the KRAS protein involved in regulating cell division.
’As people started publishing data with these antibodies that blocked EGFR it became clear that there was a good hypothesis that if you had a wild type KRAS protein you would respond to an EGFR inhibitor and if you had a mutant KRAS you wouldn’t,’ explains Tom Lillie, international head of oncology at Amgen.
After completing the Phase III study, the team revisited the samples they’d taken and established whether they had mutant or wild type KRAS genes. ’That analysis showed very clearly that it was only the wild type patients who benefitted - we didn’t see anybody with a mutant KRAS protein who had a response to panitumumab.’

One of the interesting things about the story, says Lillie, is that the science was evolving as the study was being carried out. ’When we set off nobody had really thought about KRAS as a predictive factor for panitumumab treatment. Halfway through the study a hypothesis came into being and we had to make sure we had the pieces in place to look into it.’ This is a particular problem as the pharmacogenomic field grows - biomarkers of interest often don’t emerge until later on in the drug development process, and the current clinical trial framework doesn’t easily accommodate such late stage revelations.
’When a drug is in preclinical trials you can make all sorts of assumptions about what is going to happen when you put it into humans,’ says Stephen Little, vice president of personalised healthcare at diagnostics firm, DxS, recently acquired by the sample and assay technology specialists Qiagen. ’But until the drug is being given to a reasonably sized human population, you don’t know what your response rates are going to be. It’s a big challenge.’
The Vectibix story highlights the crucial role of biomarkers in genomic medicine, and the need to be able to translate knowledge of those indicators into effective diagnostic tests that reveal whether a patient would benefit from treatment with a particular drug. Vectibix is now licensed in the US and Europe, but is associated with a companion diagnostic test that identifies which version of the KRAS gene a patient has so that only wild type KRAS patients are prescribed the drug.
Banking on biomarkers
’There are lots of different things you can measure that could be biomarkers, but they tend to fall into three classes,’ says Little; these are genetic biomarkers (like SNPs), immunohistochemistry (proteins expressed in cells) and immunoassays (looking at proteins expressed in blood). At DxS they focus on genetic biomarkers, particularly in cancers, to develop tests to be associated with certain drugs as companion diagnostics to dictate whether a patient should be prescribed the medication.
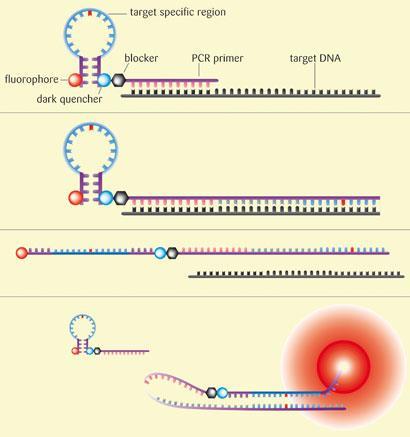
Sifting through and identifying biomarkers that could prove valuable to drug developers is a bit of a dark art, admits Little. ’There are thousands of biomarkers being published at the moment, just pulling out the one or two that look really good is quite tricky.’ But biomarkers are big business - Little says he’s not aware of a single oncology drug programme that doesn’t have a biomarker programme running alongside it. This is echoed by Lillie at Amgen - every molecule being investigated by the company’s oncology group is associated with a list of potential biomarkers early on in the development process.
’When we started this business 10 years ago, we thought the majority of biomarkers would come from looking at inherited genetic variation,’ recalls Little. ’But generally that hasn’t become as important as we thought it would, and the classes of biomarkers that have become particularly important in cancer are gene mutations and gene expression. These are dynamic biomarkers - things that change during the course of the disease and tell you something about the disease process.’
Getting at those dynamic biomarkers can be a challenge however, as you need to be able to analyse a sample, usually from a tumour biopsy. These samples are often very small, and tend to be fixed in formalin and put in paraffin blocks which can degrade the material. DxS has developed its own real-time polymerase chain reaction (PCR) technique that allows it to identify genetic mutations in these poor quality samples. ’We like real-time PCR as a technique because it’s a technology that’s well suited to dealing with the inherent variability in the samples you get from tumour biopsies,’ explains Little. The company’s system, known as Scorpions, uses molecules made of a PCR primer (a single stranded DNA fragment) covalently linked to a fluorescent DNA probe. During the PCR, the denatured target DNA sequence in the sample ’sticks’ to the primer and is replicated. After heating to denature again, if the ’new’ DNA strand is complementary to that in the probe, the two stick together and the probe fluoresces. The DNA in the probe contains the mutated sequence, and therefore the two DNA strands will only stick together, and the probe will only fluoresce, in the presence of the mutation. Using the Scorpion technology and another technique that amplifies the target DNA sequence in a sample, the company has developed a number of diagnostic tests, including screens to identify mutations in the KRAS and EGFR genes.
Biobank bonanzas
Many researchers developing companion diagnostics rely heavily on biobanks - facilities that hold vast quantities of tissue samples and associated genetic information, along with data relating to the results of drug treatments the patients may have had. Cree operates a small biobank at the oncology research centre. ’Biobanks require a lot of funding so are slightly controversial from that point of view,’ he says. ’Samples are studied to establish whether there is something that determines a good response or a bad response to treatment, and that might be predictive of that response. If it’s predictive it can be very useful. It allows you to develop companion diagnostics - doing that without access to tissue is really very difficult.’
Cree works regularly with cancer patients at his hospital, running the kind of diagnostic tests that Little develops to help determine the best courses of treatment. But he also uses the biobank and the wealth of information it holds to dig for other potential biomarkers or indicators of drug response.

Cree and his colleagues decided to look back at older chemotherapy drugs, and see if the information held within the biobank and published in the literature could reveal any correlation between gene expression and the effect of the drugs on a tumour. The team looked particularly at the most common type of lung cancer, non-small cell lung cancer, and well-established chemotherapy drugs such as docetaxel and cisplatin. The results were impressive. ’Some of the correlations we got were over 90 per cent - much higher than we had any right to expect,’ says Cree. ’And it suggests that you can predict what a drug will do in a patient by knowing what the gene expression is in the tumour. What we have got to do now is go on and prove that - and that’s where it gets very difficult.’ Moving on to develop a diagnostic that could reflect Cree’s findings would be a costly and high risk business, and as such these kinds of findings only tend to move beyond basic research if a pharmaceutical company with the money, inclination and vested interest in developing the test gets involved.
Busting the blockbuster model
Personalised medicine on a wide scale would require a complete reworking of the pharmaceutical industry’s business model. The transition from blockbuster drugs with broad target populations to more niche therapies could be uncomfortable at first - but with many pharma firms suffering as high-earning brand products lose patent protection and cheaper generic drugs take an ever growing slice of the market, it’s a leap they seem prepared to make.

And there’s money to be made for those who take the plunge - a recent report by the global consulting firm PricewaterhouseCoopers estimated the market for diagnostic tests and targeted therapies in the US alone would grow from approximately $24 billion in 2009 to $42 billion by 2015 - molecular diagnostics was singled out as ’an early winner’. The number of drugs being approved with a companion diagnostic test is also slowly growing - the FDA currently lists 19 different drugs whose labels recommend the use of a diagnostic test to select a medicine or its dose, ranging from pain killers, anti-inflammatories and blood thinners to HIV drugs, cancer therapies and antidepressants.
As the sector grows, big pharma does appear to be dipping its toes in the water. In December 2008, Swiss pharmaceutical firm Novartis waded in, setting up its own in-house molecular diagnostics unit. ’We are extending something that we have done from an R&D point of view into the market, developing companion diagnostics to support our medicines and also tests related to other disease areas we are active in,’ explains Markus Ewert, head of strategy at the unit. ’We believe that this combination of a therapy plus a diagnostic will be incredibly important in the future to achieve better outcomes for patients.’
The unit itself is a tiny proportion of the company’s business, with fewer than 100 people in a firm of almost 100,000 employees, though it works closely with a separate biomarker development group within the company. Although only 18 months old, the unit currently has nine diagnostics in its portfolio, half of which are associated with oncology.
The technological advances of recent years and lower costs are beginning to attract more players to the field, says Ewert. ’Many of the technologies that were available a few years ago in the big labs on large machines as big as a room are now becoming available in very small units that you could put in a physician’s office. Technology-wise, things are being developed so they can be cheaper, faster and more accessible.’
Newer technologies are also allowing a much broader approach to understanding disease patterns and disease mechanisms, says Ewert. ’Overall, the mindset of pharma companies has changed significantly to understand that this provides considerable additional value.’
Novartis may actually be one of the first companies to have a drug rescued from the scrap heap thanks to pharmacogenomics. Lumiracoxib, an anti-inflammatory, was approved in several countries but pulled from certain markets in 2007 following cases of severe liver damage and death in patients taking the drug. However, the company later identified a genetic biomarker that could flag patients at risk of liver damage, and in December 2009 the drug was submitted for approval in the EU in combination with the biomarker so that at-risk patients could be excluded from treatment. If the authorisation is approved, the company says it would be the first example of pharmacogenetic data being used for the reintroduction of drug withdrawn from the market for safety reasons.
Fulfilling the promise
The day when we each have a medicine cabinet stocked with drugs tailored to our specific genetic makeup is still a distant dream - and may not prove achievable or indeed practical. But the era of genomic medicine is definitely upon us, and the first strides to turn the promise of the human genome into a healthcare revolution have undoubtedly been taken.

Speaking in 2003 as the final version of the human genome was announced, Francis Collins, then director of the National Human Genome Research Institute in the US, predicted that by 2020 gene-based designer drugs would be available for diabetes, Alzheimer’s disease and hypertension, and the diagnosis and treatment of mental illness would be transformed. Halfway to that deadline, technological advances mean the $1000 genome is not far around the corner, and scientists are making good progress towards Collins’ goals, with new research on genetic determinants of disease and drug response flooding the literature. It is translating that basic research into a form that can be used in a clinical setting that is the bottleneck, and will require a rethink in the regulatory and drug development framework to ensure the gold dust discovered in the lab is not squandered, and the revolution in medical science we were promised can truly come to pass.






No comments yet