Proteins are constantly moving, but our structures of them are static. Clare Sansom talks to the researchers using free-electron lasers to make time-resolved structures
-
Serial femtosecond crystallography (SFX) allows scientists to capture proteins in motion, revealing dynamic processes like enzyme catalysis and photosynthesis in real time using ultra-short x-ray pulses from free electron lasers
-
Researchers have used SFX to study key biological systems including photosystem II and tuberculosis enzymes, offering insights into reaction mechanisms and potential drug targets
-
The technique is expensive and limited to a few global facilities, but UK scientists are advocating for a domestic XFEL to enhance access and support cutting-edge structural biology
-
While some experts are cautious about its widespread adoption, others see SFX as a transformative tool for drug discovery, enabling the design of compounds that target transient states in molecular pathways
This summary was generated by AI and checked by a human editor
Until fairly recently, everything we understood about the structure of proteins and other biological macromolecules was derived from static images. Each of the almost 200,000 x-ray crystal structure entries in the Protein Data Bank (PDB) is an atomic model of a protein or other macromolecule refined to fit experimental data. It is easy to imagine that structure as distinct and apparently unmoving. However, if you were, in a thought experiment, to shrink yourself down to protein level and look inside a living cell, everything you would see would be in constant motion.
The great 20th-century theoretical physicist Richard Feynmann once claimed that ‘everything that living things do can be understood in terms of the jigglings and wigglings of atoms’. We can certainly learn much about how proteins work from their static structures, but the complete picture will only come into focus if we explore those ‘jigglings and wigglings’ for ourselves. And this may be particularly important in the case of enzyme catalysis. Observing the process of catalytic action in real time has been a dream of enzymologists and structural biologists since at least 1965, when the very first enzyme structure – that of hen egg white lysozyme – was determined.
Plenty of those x-ray structures show the same molecule in different states or conformations, and these have been used to infer protein movements since the beginning of structural biology. Some of the unique properties of haemoglobin were deduced from small structural differences between the ‘tense’ and ‘relaxed’ forms of the protein with, respectively, low and high oxygen affinity.
It has taken a step change in the methodology of x-ray crystallography possibly equivalent to the birth of film from still photography to make it possible to deduce function from molecular movements across a time series of ‘stop-motion’ atomic models progressing along a reaction coordinate.
Diffraction before destruction
In conventional x-ray crystallography, a single, largish crystal (typically at least 0.1mm in one or more dimensions) is held at very low temperature (–173°C) and rotated in a beam of x-rays to collect enough data to obtain a 3D structure. In contrast, serial femtosecond crystallography (SFX) involves passing a stream of much smaller crystals across a pulsed beam of x-rays. Each pulse is powerful enough to cut through steel, but short enough to obtain an x-ray diffraction pattern from a tiny crystal in the fraction of a second before its target – not surprisingly – explodes. This technique, which has been called ‘diffraction before destruction’, has many advantages for structural biology. It can be carried out at room temperature; the pulses are short enough to outrun the radiation damage that occurs when x-ray beams hit delicate protein crystals; and, crucially, the short pulses freeze the molecular motion like a strobe light. Consequently, if scientists collect enough data at a number of timepoints to solve atomic models related in time, then they can string together the series of strobed structures to create a ‘stop-motion movie’ of enzyme catalysis.
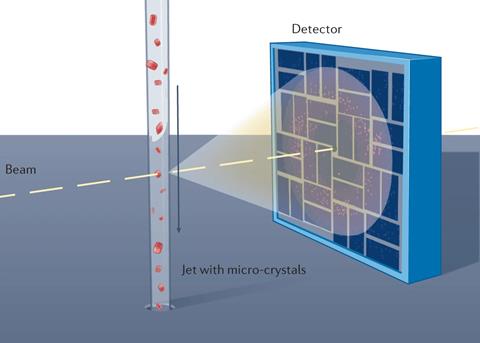
The pulsed beam of intense x-rays used in SFX is produced by a x-ray free electron laser (XFEL) in which electrons are accelerated close to the speed of light and passed through a series of magnets known as an undulator to constantly change their direction. This generates a high-intensity beam of electromagnetic radiation, which can be tuned to any frequency from microwaves to x-rays. The free electron laser itself was invented in 1971 but has been used in structural biology for under two decades. The British-born physicist Henry Chapman, then at DESY in Hamburg, Germany, led the large international collaboration that first demonstrated that the technique could be used for observing protein structures in the early 2010s. He was awarded the 2015 Leibniz Prize by the German Research Foundation for this pioneering work.
The technique requires, besides a source of intense x-ray pulses, a steady stream of microcrystals. Several methods of sample delivery have been developed, and most are also compatible with a variety of reaction initiation strategies. Allen Orville, now group leader of the XFEL Hub at Diamond Light Source in the UK, explains: ‘I came across Chapman’s papers when working at Brookhaven National Lab (BNL) in New York, and that changed the course of my research career. I developed a proposal for a kind of ink-jet printer to launch nanolitre-scale, crystal-containing droplets into an x-ray beam intended for NSLS-II at BNL. We took our prototype to the LCLS XFEL at Stanford, California, but although we launched droplets from the well meniscus into the x-ray pulse, it failed to deliver crystals because they all sank to the bottom of the well. So, we decided to work with gravity rather than against it. We turned the instrument upside down and started launching the droplets downwards; that worked first time, and we still use the technique at XFELs today.’
An XFEL is extremely expensive, and there are still very few facilities worldwide. The Lightsources.org collaboration includes eight in continental Europe, the US, Japan and South Korea. Chapman is now a director at the Center for Free-Electron Laser Science (CFEL), also in Hamburg. This is co-located with the electron gun for the world’s largest and most powerful x-ray laser, the European XFEL. Its electrons are generated at DESY, accelerated in a linear particle accelerator and rapidly reach speeds close to that of light; the beams pass through the series of undulators and come to an end in an experiment hall in the small town of Schenefeld, 3.4 km away . ‘The high power of our x-ray laser means that our data will have a high signal-to-noise ratio, and the structures we determine will have high spatial as well as temporal resolution,’ says Sakura Pascarelli, European XFEL scientific director.
Watching photosynthesis – in real time
To use SFX – or, indeed, any other technique – to follow a reaction such as an enzyme catalysis in ‘real time’, that reaction must be triggered. Practically, the easiest such trigger to use is a pulse of visible laser light, so it is not surprising that many of the structural biology papers to have come out of XFELs so far follow the reactions of photosensitive proteins. ‘One of the first proteins to be studied in detail by SFX, and an ideal system to start with, was photosystem II, a protein complex involved in photosynthesis,’ explains Ilme Schlichting, a director at the Max-Planck Institute for Medical Research in Heidelberg.
Photosynthesis is the process through which plants, algae and some bacteria convert light energy from the sun into chemical energy, and it produces molecular oxygen as a byproduct. Understanding the complete mechanism of action for photosystem II (PS II) at a molecular level has been described as a ‘holy grail of structural biology’, and following the catalytic mechanism of photosystem II in real time has taken us a few steps closer to this particular grail.
Despite its name, photosystem II is the first complex to be activated when sunlight strikes a leaf. It is a large protein, a dimer in which each monomer consists of 20 different protein chains, and it sits in the thylakoid membrane within the chloroplast of a plant cell. In simple terms, it harvests light photons that energise its electrons; each electron passes along a chain through the protein between coenzymes and cofactors until it reaches a molecule of plastoquinone, which is reduced to plastoquinol. Water is oxidised to molecular oxygen and ionic hydrogen, thus replacing the electron lost to the plastoquinol, and the oxygen molecule is released. ‘Conventional crystallography using a continuous x-ray beam can only give the structure of the complex in its dark, inactive state,’ says Jian-Ren Shen of Okayama University, Japan. ‘The activated complex passes through its intermediate states very quickly, and you can only observe these if you use very short, intense x-ray pulses of perhaps 10 femtoseconds.’
Shen is using the SACLA XFEL in Japan to follow the photosystem’s transitions between intermediate states and observe the mechanism through which the double bond between the atoms of molecular oxygen is formed. ‘Illuminating the stable, inactive “dark” complex, which is in a conformation termed S1, with either one or two flashes of visible laser light initiates a very rapid transition to the unstable S2 or S3 state, and we can watch the structure changing continuously by collecting XFEL data at different time points afterwards,’ he explains. The bond-forming reaction occurs at the oxygen-evolving complex, which is a Mn4CaO5 cluster embedded in the photosystem. The most significant observed change occurs after the second light flash, when a water molecule disappears from the electron density map and a new oxygen atom is seen very close to another in the cluster. As the cycle progresses the distance between those two oxygens decreases from about 2Å to something very close to the bond length of molecular oxygen, 1.2Å.
‘We now intend to solve the final structure of the photosystem, after the oxygen molecule is released,’ says Shen. ‘It is very difficult to see this structure using serial crystallography, because every time light induces a change of state, some molecules are “left behind” and we end up with a superposition of states. Instead, we illuminate the protein so it goes through the whole cycle, freeze it, and then solve the structure using cryo-electron microscopy.’ This is a good example of the synergy that can occur between the two techniques.
Tuberculosis in the spotlight
Serial femtosecond crystallography also has a role to play in combatting the intractable problem of antibiotic resistance. For almost a century, the discovery and development of antibiotics has relied on the fact that bacterial enzymes differ in function, or at least in structure and mechanism, from anything found in the human proteome. The more we can learn about those structures and mechanisms in molecular detail, the easier it will be to keep ahead of the constantly evolving bacteria, and this technique adds a fourth dimension – time – to three-dimensional molecular structures. In these cases, however, the reaction to be followed is generally triggered not by light but by the addition of a ligand such as the substrate of an enzyme.
The bacterium Mycobacterium tuberculosis may be responsible for more human deaths throughout history than any other pathogen, and it is still a killer: about 1.25 million people lost their lives to tuberculosis in 2023 alone. While drug-sensitive TB can be controlled relatively easily, multi-drug resistant and extremely-drug resistant forms are increasingly prevalent, and new drug targets, and new drugs for established targets, are increasingly needed. Chapman’s group has been part of a large collaboration studying the enzyme that inactivates penicillins, beta-lactamase, and its inhibitors in M. tuberculosis. ‘We have seen how the inhibitor approaches the active site and its induced fit, and we are increasing the spatial and temporal resolution of the structures,’ he says.
Furthermore, M. tuberculosis contains the common enzyme alanine racemase, which catalyses the conversion between the L- and D- forms of the amino acid alanine. This is a good drug target that has been rarely used in the clinic. It is, however, one of the targets of cycloserine, an antibiotic that is primarily used for treating drug-resistant tuberculosis. Several models had been proposed for its reaction mechanism before Ki Hyun Nam, Yunje Cho and their colleagues probed its dynamics using the XFEL in Pohang, South Korea. This suggested a pathway for the substrate through the active site that should prove useful in the design of further inhibitors.
From femtoseconds to attoseconds
Serial femtosecond crystallography is suitable for studying the molecular detail of processes like these because atoms typically move, and proteins change conformation, in timescales no faster than femtoseconds. This, however, is still too slow to follow the movement of electrons and, for example, to see bonds form and break in real time. For that, we need to measure time in attoseconds (1as = 10–18s), and this incredible feat is becoming possible. The 2023 Nobel prize for physics was awarded to Anne L’Huillier, Pierre Agostini and Ferenc Krausz for demonstrating that it is possible to create attosecond-length pulses of light. As Jon Marangos, a physicist and ‘attosecond scientist’ at Imperial College London in the UK, explains, ‘one attosecond is to one second as one second is to the age of the universe’. Scientists have now demonstrated x-ray pulses as short as a few hundred attoseconds, although these have not yet been employed in structural biology.
The UK is one of the 12 European countries that run European XFEL, and its scientists have ready access to other European XFELs, but there is not yet one in the UK. The UK’s Science and Technology Facilities Council has put together a consortium to develop a detailed science case and a conceptual design for constructing one. They argue that it would ‘enhance the international portfolio of XFELs, offer the UK’s structural scientists in all disciplines a step change in capability… and contribute to solving some of the world’s most pressing scientific problems’.
Marangos has taken the lead on developing this case, with Orville leading on the life sciences section of the proposal and particularly on its use for time-resolved structural biology. ‘Even putting the case together is a risky, long-term project,’ explains Marangos. ‘We began it before the pandemic and finished the first edition during lockdown, and now five years on we must update it to include the recent developments using XFELs across many fields … we still have to decide on the most appropriate scale, optimise the design for high throughput to increase cost effectiveness, consider alternative options and location for any UK facility.’ There is no guarantee that the project will go ahead and even if it does, and a design is signed off within the next year or two, no UK XFEL could generate data until around 2035. The UK XFEL Hub’s other task, to help its scientists access free electron lasers elsewhere, will be crucial for some time to come.
The technique is unique for studying reactions on an ultrafast timescale
Time-resolved structural biology may be the most common application of free electron laser technology in the life sciences, but it is not the only one. Colin Berry of Cardiff University in Wales has found the technology appropriate for his studies of bacterial proteins that are used in mosquito control. ‘These proteins crystallise naturally in inclusions within the bacteria, but the crystals are too small to be useful in conventional crystallography,’ he explains. ‘Before XFEL came along we tried and failed to get them to form larger crystals, but this technique allows us to obtain high-resolution structures from the natural nanocrystals without re-crystallising.’ Understanding the basis of ligand binding and the mechanism of action of these proteins, which are toxic to the mosquito vectors of diseases such as elephantiasis and West Nile fever, could greatly enhance their usefulness as biocontrol agents.
What does the future hold for serial femtosecond crystallography? Will time resolved crystallography and ‘diffraction before destruction’ take over from conventional crystallography as electron microscopy also expands its reach, or will it stay in a niche? Schlichting is among the pessimists. ‘Structural biology applications at XFELs will remain limited,’ she says. ‘The technique is unique for studying reactions on an ultrafast timescale, for example establishing energy flow through a protein and inducing structural changes, but it is unlikely to become part of a generic structural biology toolbox. Less complex alternative methods are available for time-resolved studies of protein structure, including hydrogen–deuterium exchange.’
Orville is more optimistic about the future of the serial crystallography technique and notes many emerging XFEL-like applications at synchrotrons too, which greatly increases access by many more scientists. He is also particularly keen on potential applications in early-stage drug discovery. ‘Most often, in drug design, people look for an inhibitor that binds to the ground state of the target enzyme,’ he says. ‘With the right tools [SFX] we’ll be able to find compounds that bind to any point on the reaction pathway … and that will provide extra specificity.’ He often names Vertex’s new sodium pump inhibitor, suzetrigine, as a ‘poster child’ for the potential of the technique. ‘This drug, which the FDA approved in January 2025, is the first truly novel analgesic to be licensed for over 20 years,’ he explains. ‘Understanding the dynamics of the ion channel was essential for its design, and, more generally, this proves that drugging the dynamics of a system is not far-fetched anymore.’ Time will tell which of these futures will come to pass, but my money is on Orville’s vision of molecular movies aiding drug design.
Clare Sansom is a science writer based in Cambridge, UK














No comments yet