New sensor system uses gold nanoparticles to detect proteins indicative of disease in human blood serum
US scientists have developed a sensor system for profiling the protein content of human blood serum without needing individual receptors for each separate protein. The technique could be used to provide cheap preliminary disease diagnosis information, saving time and money by reducing unnecessary testing.
There is a direct link between levels of protein in human serum and disease, making protein levels an attractive target for sensors and diagnostics. However, with over 20,000 proteins present in human serum at a range of different concentrations, developing an effective sensor has been a challenging task.
Pattern recognition sensing uses detector systems that interact with many different proteins to give different responses. These responses together help build up an overall diagnostic signal pattern in much the same way as the human nose works, taking patterns of inputs from a few hundred types of receptors to identify many thousands of different ’smells’.
’People have done "chemical nose" sensing of proteins before,’ explains Vince Rotello from the University of Massachusetts in Amherst, US, ’but they’ve always been in buffer. Taking it into real world [blood] serum makes life a lot more difficult, but also gets us a lot closer to a clinically useful system.’
Techniques to detect specific proteins can be expensive because they generally use individual antibodies or complex mass spectrometry techniques and are limited to detecting only their target protein. Rotello explains that they needed a more general receptor system. The team used gold nanoparticles coated with short polymer chains that can be altered to tune the surface chemistry of the particles to favour binding to different types of proteins.
’That’s the recognition part,’ says Rotello, ’but you also need to generate a signal, and for that we used Green Fluorescent Protein (GFP).’ The GFP sticks to the nanoparticles, which quenches its fluorescence, explains Rotello. ’When it’s kicked off the particles by a competing protein, it lights up,’ he adds. Different proteins will compete more or less strongly with GFP to bind nanoparticles with different surface chemistry, so the way a serum sample interacts with several different nanoparticles gives a fluorescence profile that reflects the concentrations of the proteins.
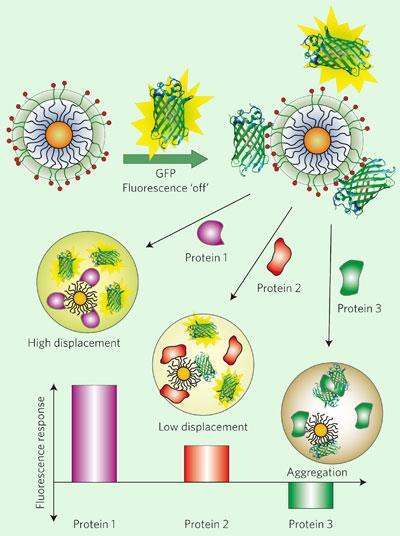
Using GFP as the reporter molecule will really help the technique be accepted by other researchers, says Eric Anslyn from the University of Texas, Austin. ’GFP is a really standard biochemical tool [as recognised by the Nobel prize for Chemistry in 2008], so molecular biologists will feel very comfortable using it.’
However, just as we have to learn to recognise smells, the team had to teach their system to distinguish the different proteins in blood serum. There are more than 20,000 different proteins in serum, but over 90 per cent of the total protein content comprises just five proteins, so the team initially focused on discerning these five. They found a set of three nanoparticles that work well to distinguish the proteins, and trained the system by varying the concentrations of one at a time. ’One of the things we’ve shown is that we don’t need to test every concentration to generate a useful pattern,’ says Rotello, ’which is important because the idea of having to combinatorially vary every protein in serum becomes impossible.’
The working system can detect variations in concentration as small as 500 nM for any of the five proteins - even human serum albumin (HSA), which makes up 70 per cent of the total protein count. Rotello was surprised by how sensitive it was: ’It’s like there’s a haystack of HSA, and we’re seeing addition of couple of extra bits of hay on the top, but we can also see the needles inside.’
Anslyn is very enthusiastic about the research: ’These nanoparticles are brilliant for making receptors for surface recognition of proteins,’ he says, ’and now they’ve shown that their arrays work even with the background of all the other proteins in human serum - no-one else has done that yet.’
Rotello hopes that the system can be refined to give quick and cheap indicative profiling of proteins in human serum. The system could recognise the patterns caused by protein imbalances in various diseases, ’I don’t see it giving definitive answers,’ he says, ’it tells you there’s something wrong and gives some diagnostic information to work with.’
Phillip Broadwith
References
et alNature Chem, 2009, DOI:10.1038/nchem.334


No comments yet