Researchers stumble upon biochemical machinery that helps bacteria make essential B vitamin
US researchers have revealed the identity of an enzyme1 used by bacteria to make the essential B vitamin folate, 30 years after it was first isolated. The team, led by Mario Amzel and Maurice Bessman of Johns Hopkins University, Baltimore, Maryland, say the enzyme could be a potential target for antibacterial drugs.
When bound to enzymes in the body, each folate molecule contributes a single carbon atom to the manufacture of important biological compounds, including DNA during cell division.
Although humans cannot synthesise the vitamin, relying instead on their diet to provide sufficient folate, bacteria can produce it for themselves. This makes the biochemical synthesis of folate an ideal target for antibiotics - knock it out and, hopefully, patients will not suffer too many side effects. As a result, researchers have studied the pathway intensively, making the find all the more surprising.
’There are very few opportunities to discover a pathway enzyme these days,’ Amzel told Chemistry World. ’But it seems there are still a few holes here or there.’
The team was studying an enzyme produced by the orf17 gene in E.Coli, known to catalyse the hydrolysis of deoxyadenosine triphosphate (dATP) to deoxyadenosine monophosphate (dAMP) and pyrophosphate.2 But it was only when they chanced on a paper3 published in 1974 by Yuzuru Suzuki and Gene Brown of the Massachusetts Institute of Technology, Cambridge, US, that they realised this enzyme might also catalyses the second step in the folate pathway. Suzuki and Brown had 30 years earlier isolated the enzyme, which converts dihydroneopterin triphosphate (DHNTP) to dihydroneopterin triphosphate (DHNMP) plus pyrophosphate, but were unable to identify the gene coding for it.
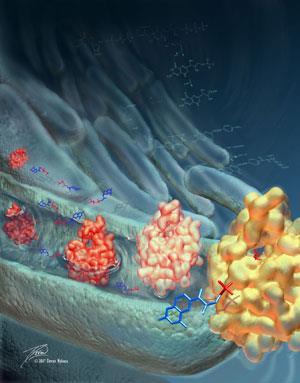
The Johns Hopkins team found their enzyme hydrolysed DHNTP seven to eight times faster than dATP. Deleting the orf17 gene reduced the amount of folate the bacteria were able to produce ten-fold, confirming they had found the last missing piece of the folate synthesis pathway. They were also able to track the enzyme down in other species of bacteria.
Amzel says the enzyme is another potential antibacterial target. ’We are not in the business of drug design . [but] we do hope that others can use what we find to make new drugs.’
The researchers also found the protein, which is a member of a large family of related enzymes known as the Nudix hydrolases, may have an unusual catalytic mechanism. While most family members use a glutamate as a catalytic base in the reaction, in this enzyme it is most likely a tyrosine, says Amzel.
Enzymologist Alexander McLennan of the University of Liverpool, UK, says if that is confirmed by further experiments it would make the Nudix family unusual if not unique.
’The family may be using different catalytic bases, which is strange,’ McLennan said.
But he doubts the newly discovered enzyme will be useful for drug designers because blocking it merely slows bacteria down, rather than killing them outright.
’The folate pathway is certainly very important as an antibiotic target,’ McLennan said. ’But this particular enzyme would not be my target of choice within it.’
Ananyo Bhattacharya
References
et al, Structure15et al,J. Biol. Chem., 273J. Biol. Chem249, 2405





No comments yet