Tony Campbell's fascination with 'living light' - the bioluminescence responsible for the glowing colours of fireflies, glow-worms and jelly fish - has led him to develop a range of colourful proteins.
Tony Campbell’s fascination with ’living light’ - the bioluminescence responsible for the glowing colours of fireflies, glow-worms and jelly fish - has led him to develop a range of colourful proteins that light up along the same principles
Twenty years ago I and my coworkers invented a technology that has revolutionised a major area of clinical diagnosis. This involved replacing radioactive iodine used in immunoassay by an acridinium ester that makes its own light - chemiluminescence. This type of immunoassay is now used in 100 million clinical tests every year worldwide, allowing clinicians to measure a wide range of proteins, pathogens and other molecules in blood samples.
The original idea for the technology all started with my curiosity about how bioluminescent animals can produce ’cold’ light, and how they flash or glow (see Box). I then became curious about how these creatures generate different colours. I saw that Nature is capable of generating a complete rainbow of colours. This later led me to realise that we can mimic what Nature has achieved over millions of years of evolution, and generate a new technology, Rainbow proteins, with even greater potential than immunoassay alone.
Rainbow proteins could revolutionise drug discovery and direct diagnosis in a GP’s surgery or at a patient’s bedside, called point-of-care testing. The significance of Rainbow proteins is that they demonstrate exquisite sensitivity coupled with homogeneous assay, ie all the reagents are added with the sample in the same tube. Bioluminescent probes can be detected down to just a few hundred molecules in a swimming pool of water - impossible with fluorescence. As a result they can solve many of the current problems in the diagnosis of diseases such as cancer and heart disease, and in high throughput drug screens.
So where did it all begin? My interest in bioluminescence began when I was a PhD student at the University of Cambridge. I used the chemical reaction that makes fireflies flash and glow-worms glow to measure tiny amounts of ATP - the energy currency of life. I was then lucky enough to be introduced to the Marine Biological Association Laboratory at Plymouth. There I discovered a luminous hydroid called Obelia, part of the life cycle of a small jelly fish. It produces a protein that flashes in the presence of calcium, thereby allowing me to measure free calcium inside live cells. This turns out to be a universal chemical switch making muscle contract, an insulin cell to secrete, and so on.
In 1979 I was invited to join a research cruise on RRS Discovery to explore the largest ecosystem on our planet: the deep sea. Here I found an incredible diversity of animals that make their own light. And we observed the most common chemical responsible for bioluminescence in the sea, called coelenterazine (1) after the coelenterate jelly fish in which it was first discovered. As a result of studying bioluminescence in these marine creatures and land animals like glow-worms, I found that there are essentially three ways by which Nature generates its rainbow (see Fig below):
1. Animals such as fireflies generate light via the oxidation of a small molecule called a luciferin, catalysed by a luciferase enzyme. Different luciferins produce different colours. So, for example, the luciferin in luminous beetles produces green, yellow or red light. But marine creatures that use the most common luciferin in the sea emit violet, blue or green light.
Luciferase
Luciferin + O2 + cofactors ___________> oxyluciferin + light
2. The environment created by the luciferase protein when it binds to the luciferin during this process also affects the colour of the emitted light. In fact all the luciferase does is create a solvent cage around the luciferin, allowing one and only one oxygen atom in, and keeping water out (both oxygen and water quench electronically excited states).
3. The transfer of energy from a donor molecule in one protein to an acceptor in another is another way of generating bioluminescence. The most famous example of this is the transfer of energy from a normally blue emitting photoprotein or luciferase to a fluorescent protein called green fluorescent protein (GFP), resulting in the emission of green light.
Three key luciferins in bioluminescence, and the three types of Rainbow proteins
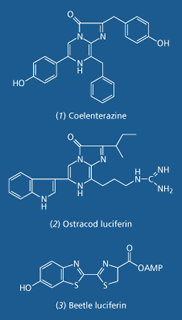

The most common compound responsible for bioluminescence in the sea, coelenterazine, is a type of imidazolopyrazine. Coelenterazine is the luciferin in luminous organisms in seven distinct phyla, while a similar imidazolopyrazine (2) is the luciferin in some ostracods (water-loving crustaceans) and fish. Imidazolopyrazines emit violet to blue-green light.
But coelenterazine has never been found in terrestrial luminous animals. In the thousand or so luminous beetles the luciferin is a benzothiazole (3), which is responsible for the emission of green, yellow or red light. Red bioluminescence is surprisingly rare, being found only in the beetles, and a group of deep sea fish, the dragon fish.
In the late 1980s I realised that if we could get the messenger RNA or DNA that codes for these proteins into live cells, human cells for example, then the light emitted would allow us to measure chemical reactions inside the cells, and even in separate cellular compartments. Thus we made the first type of Rainbow protein, using two separate chemistries, yellow emitting firefly luciferase linked to blue emitting aequorin, measuring ATP from yellow light and Ca2+ from blue light.
But the precise colour emitted by each luciferin depends on the environment created by the luciferase. This led at the same time to the generation of the second type of Rainbow protein. The presence of the enzyme can affect the electrochemistry, for example the pH inside the luciferase affects the ratio of different ionised states of the luciferin, which in turn produce different colours. Alternatively, the luciferase affects the conformation of the luciferin. One conformation can produce green light, the other red light.
But the crucial discovery for me came in the late 1980s when I and my colleagues at the University of Wales College of Medicine started to look at the exact protein sequences of the luciferases. When a different luciferin is involved the luciferases are completely different. There is no similarity in their amino acid sequences. In contrast, when the organisms are in the same group the luciferase can be remarkably similar. We found that the green emitting British glow-worm, Lampyris noctiluca, has a luciferase that is 80 per cent identical to that of the yellow emitting American firefly, Photinus pyralis. Just one or two amino acids are responsible for the change of colour that occurs during oxidation of the same luciferin compound.
This observation set me thinking. What if we engineered other amino acids into the luciferase that would react with substances - say other enzymes such as kinases or proteases - that we wanted to measure inside live cells? Surely when the resulting enzyme reacted with these substances there would be a change in colour and/or light intensity? This is the ’rainbow’ effect. And it worked.
Thus in 1991, we were able to measure protein kinase A in a live cell, or a key protease in blood clotting - thrombin. We found two places in firefly and glow-worm luciferase where we could engineer sites for protein kinase without inactivating the enzyme. One was near the luciferase active site, the other was at the end of the protein - the C-terminus. The last 12 amino acids of the luciferase could be replaced by active site sequences from a wide variety of peptides, including those with kinase and protease sites.
I developed the third type of Rainbow protein, based on energy transfer, in the mid-1990s. The original idea had come from my early work with the hydroid Obelia while at Plymouth in the 1970s. Hydroids look at first glance like small plants, but are actually colonies of animals with sting cells. Thousands of Obelia appear on brown seaweed in the summer, and each generates dozens of small jelly fish. The live Obelia and its constituent jelly fish emit blue-green light with a peak wavelength of about 508nm. But when I extracted the protein thought to be responsible, this emitted blue light with a peak at 475nm. I called this protein obelin.
Like its American relative aequorin, obelin’s luciferin and oxygen are so tightly bound to the protein that the three can be isolated together as one complex. All you have to do is add calcium and the complex flashes. It is thus named a photoprotein. But the reason for the difference in colour between the animal and the extracted photoprotein turned out to be GFP, which accepts the energy from the excited oxyluciferin before it has a chance to release a photon. GFP then emits the photon, now green.
Whilst cleaning up at Plymouth late one night I had a flash of inspiration. What if we could engineer the two proteins together with a reactive site in between, or bring them together in an antibody-antigen complex. Surely when a substance I wanted to measure, say an antibody or protease, reacts with this site it would change the colour of the light - what I now call the rainbow effect. At the time, 1978, cloning was a dream of science fiction. By the mid-1990s we had the respective DNA sequences for each protein and we were able to show that it worked.
Thus engineering GFP to aequorin with a peptide linker in between resulted in complete transfer of energy from the excited oxyluciferin to the light-emitting ’fluor’ moiety of GFP. This emits green light. But once the linker is cleaved in the presence of a protease enzyme called caspase, energy transfer is no longer possible. So the light returns to blue. This allowed us to detect the activation of caspase, the first enzyme in the cell death (apoptosis) pathway, in a live cell. Finding such a ’blue needle’ in a ’green haystack’ has huge potential in screening for new drugs.
Lighting up the chemistry of living cells. The endoplasmic reticulum lit up by GFP (left), and (right) a flashing luminous leaf. This picture was taken by a special camera that images individual photons. The light is captured by an array of 250 000 pixels, seen as the tiny squares that make up the resulting picture. The computer converts the light intensity in each pixel to a colour: blue is dim, red is bright.


A further breakthrough was learning how to target our Rainbow proteins to specific sites within cells. In nature, different proteins come with specific peptide tags that act as address labels to direct the molecules to specific sites within a cell. By similarly tagging our Rainbow proteins we are able to measure Ca2+, ATP and enzymes in organelles such as the endoplasmic reticulum (ER), mitochondria and nucleus, and on the inner surface of the plasma membrane.
So what have we found out? Bioluminescent proteins, together with fluorescence and molecular imaging, have revolutionised our understanding of cell biology. Free Ca2+ has been identified as a universal signal for many cell events - for all forms of muscle contractions and the secretion of insulin for example. This works because there is a huge gradient of free calcium across the membrane of all living cells. Outside the cell the calcium level may be 1-10mM, but inside the free calcium concentration can be as low as 0.1?M. Because of this ’calcium pressure’ a tiny movement of calcium into the cell, or release from an internal store, causes a very large-fold increase in free calcium inside the cell. In some cases, after a stimulus such as a neurotransmitter or hormone has hit the outside of the cell, the level of free Ca2+ inside can rise a hundred times or more. It is purpose-made to be a switch. We can even watch calcium switches now in live bacteria and whole plants.
Rainbow proteins are the next generation of luminescence technology that I began in the 1970s. They can also light up the human mind. We are using bioluminescence in our Darwin initiative in Pembrokeshire to promote science education and the public understanding of science. And it all started by me being curious about how animals that make their own light can produce a rainbow.
Source: Chemistry in Britain
Acknowledgements
I thank my wife, Stephanie Matthews, and family for their support. Thanks also to all the people in my group over the past 30 years, Howard Potter and David Burton for helping to set up AKRainbow, and those who helped make the the Darwin Centre in Pembrokeshire a reality. This work was supported by funds from the MRC, NERC and Wellcome Trust.
Anthony K. Campbell.
Further Reading
- A. K. Campbell, Chemiluminescence: principles and applications in biology and medicine, p608. Chichester and Weinheim: Horwood/VCH, 1988.
- A. K. Campbell, Rubicon: the fifth dimension of biology, p304. London: Duckworth,1994.
- A. K. Campbell and S. B. Matthews, Lactose intolerance and the MATHS syndrome: what are they and how can I cope?, p32. Pembrokeshire: Welston Press, 2001.
- A. K. Campbell, A. J. Trewavas and M. R. Knight, Cell Calcium, 1996, 19, 211.
- S. Dunstan et al, J. Biol. Chem., 2000, 275, 9403.
- J. M. Kendall et al, Biochem J., 1996, 318, 383.
- J. W. Hastings and T. Wilson, Annu. Rev. Cell Dev. Biol., 1998, 14, 197.
- G. B. Sala-Newby et al, Meth. Enzymol., 2000, 305, 478.
- C. M. Thomson, P. J. Herring and A. K. Campbell, J. Biolum. Chemilum., 1997, 12, 87.
- J. Waud et al, Biochem. J., 2001, 357, 687.
- J. Waud, G. B. Sala-Newby and A. K. Campbell, Biochim. Biophys. Acta, 1996, 1292, 89.
Living lights - bioluminescence
Bioluminescence, living light, is the emission of light from living organisms. Bioluminescence is found in a wide range of microorganisms, fungi, invertebrates and fish, and is particularly common in the deep sea, where virtually all creatures make light. These organisms emit light over a whole rainbow of colours, from deep violet, through cyan, blue, green, orange to red. All bioluminescence is ’cold light’, generated as the result of a chemical reaction (chemiluminescence). When a candle burns, the chemical reaction generates heat, which in turn produces incandescence, and thus light. But in bioluminescence, a special type of chemiluminescence, all the energy from the reaction, the enthalpy, goes to make light. The reaction is still ’burning’ - oxidation is occurring - but no heat or ’fire’ is being produced.
Eilhardt Weidemann was the first to use the term luminescence in 1888, some nine years before J.J. Thompson reported the discovery of the electron. We now know that all types of luminescence require an electron to be excited. Different types of luminescence use a particular energy source for excitation. Photoluminescence (fluorescence and true phosphorescence) requires the absorption of visible or UV light, or infrared in the case of multi-photon excitation. Triboluminescence requires major structural changes between molecules, such as fractures, eg a peppermint being cracked, or two quartz crystals being knocked together. But in chemiluminescence the energy required to excite the electron comes from a chemical reaction. The secret in chemiluminescence is that the key reaction intermediate must generate its energy very rapidly so that the electron can be excited before the chemical energy is lost as heat.

There are two common mechanisms for this: radical annihilation, and the cleavage of a four-ringed structure called a dioxetane. Radical annihilation occurs in the upper atmosphere, whereas dioxetanes (see Fig above) are the major chemical source of bioluminescence. This last mechanism involves two carbons linked together, bridged by two linked oxygens. In some bioluminescent reactions this highly unstable intermediate is in the form of a dioxetanone where one of the carbons is in the form of a carbonyl. Spontaneous cleavage of the bond between the two carbons generates two carbonyls, and there is enough energy in this cleavage to excite one of them electronically. Light is then emitted when the electron returns to the ground state.
In 1947 Bill McElroy at Johns Hopkins University in the US showed that ATP (the energy currency of cells) was required for the chemiluminescent reaction in fireflies, and all other luminous beetles. But he got the right answer for the wrong reason. ATP hydrolysis generates less than 10kcal mol-1. To generate a visible photon you need 50-100kcal mol-1. Thus ATP is not the direct energy source for any bioluminescence. It allows the reaction in the firefly to occur quickly enough to generate all the oxyluciferin molecule in an excited state. But the energy for the light still comes from ’burning without fire’.
In bioluminescence the signal for light emission is either the mixing of the key three components (luciferin, luciferase and oxygen), or the generation of a trigger by the luminous cells. For example, in fireflies and glow-worms it is access to oxygen that allows the beetle to flash or glow. In contrast, in luminous jelly fish calcium penetrates the cell, binds to the key protein, and triggers it to flash. This means that each bioluminescent reaction can be used to measure one of the components of the reaction or the trigger. Thus measurement of free calcium in cells using the jelly fish or hydroid protein has identified this humble cation as the crucial signal of many cellular processes, including all types of muscle contraction and secretion. Every second your heart cells are generating a tiny calcium puff to allow your heart to beat and keep you alive.
The luciferase from fireflies is the standard assay for ATP. Since all living cells have a lot of ATP, and dead cells have very little, ATP is a very sensitive way of detecting biomass, the contamination of food with live bacteria for example. As little as 10-17mol (10 attomol) ATP can be detected. A photoprotein called pholasin can be used to measure the generation of toxic oxygen species, superoxide, by a single human neutrophil (a type of white blood cell), vital to understanding crippling diseases such as rheumatoid arthritis. Pholasin is found in a bivalve known as the piddock (Pholas dactylus). Whilst studying the hydroid Obelia I could detect as little as 10-21mol of its luminous protein obelin in a swimming pool of water. Once we had the DNA from animals such as Obelia, glow-worms and fireflies. I realised that this DNA could be used to generate the luminous indicators in live cells, and even intact organisms.
Firefly luciferase or GFP DNA are now used as very sensitive markers for detecting the activation of specific genes in living cells. This is done by engineering the DNA coding for firefly luciferase or GFP on to the element that controls the switching on and off of the particular gene being studied. When this element is switched on the cell will light up yellow or green. GFP also allows us to light up structures and even the movement of specific proteins in single living cells. In the picture below the endoplasmic reticulum is lit up by GFP linked to a protein called calreticulin that is a crucial signalling protein inside this structure. Bioluminescent indicators really have revolutionised cell biology by lighting up the chemistry of living cells.
Contact and Further Information
Anthony K. Campbell
Professor in medical biochemistry
Department of medical biochemistry, University of Wales College of Medicine, Heath Park, Cardiff CF14 4XN
Tel: 02920742951
Fax: 02920745440
Anthony K. Campbell
Scientific advisor
AKRainbow


No comments yet