New antibiotics designed with the help of AI show promise against two deadly drug-resistant bacteria. Two of the new compounds attack bacteria in ways unseen in existing drugs.
Development of new antibiotics has failed to keep pace with rising antibiotic resistance in bacteria, leading to an urgent public health crisis. Once-treatable diseases are turning into relentless foes, with antibiotic-resistant bacteria linked to approximately five million deaths each year.
Now, biomedical engineer James Collins from the Massachusetts Institute of Technology, US, and his team have used a deep learning model to design entirely new antibiotics. Using a graph neural network that links chemical structure to predicted properties, the researchers sifted through more than 45 million chemical fragments to identify structures with potential antibacterial activity against Neisseria gonorrhoeae and Staphylococcus aureus.
After manually filtering out fragments with undesirable properties like toxicity, metabolic instability and promiscuous binding, Collins’ team used a subset of promising fragments to act as seeds for two generative AI models. These models specialised in fragment-based design, in which molecules are created by modifying different side-chains, and de novo design, in which molecules are generated from scratch. The AI models designed over 36 million compounds and screened them for their potential antibacterial activity.
Highly potent
Collins’ team then synthesised 24 of the AI-generated compounds, with two molecules, called NG1 and DN1, standing out due to their potency, selectivity and entirely new mechanisms of action.
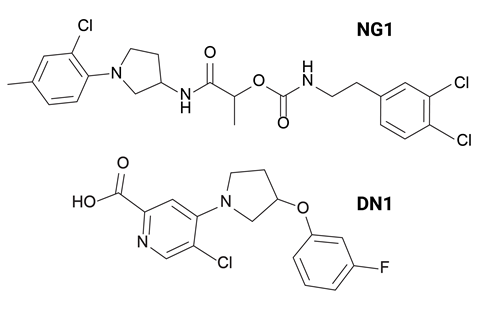
‘NG1 selectively targets pathogenic N. gonorrhoeae’ while sparing beneficial commensal Neisseria species such as N. cinerea and N. mucosa, which are important for maintaining a healthy vaginal microbiome,’ says Aarti Krishnan, who worked on the project. ‘It also killed the highly drug-resistant strain found in the US that lost susceptibility not just to ceftriaxone but to all other drugs previously recommended for first-line treatment.’
DN1 showed strong bactericidal activity against S. aureus with its potency comparable to those of other frontline antibiotics like vancomycin and linezolid. Its bactericidal activity was rapid, notes Krishnan, ‘achieving complete killing within four hours, whereas vancomycin requires around ten hours’. The compound was also active against strains that are resistant to current antibiotics.
Jonathan Stokes, a computational chemist at McMaster University in Canada who wasn’t involved in the work, calls it an ‘interesting demonstration’ of generative AI’s use in early-phase antibiotic discovery, ‘particularly in exploring bioactive chemical space against two distinct pathogens’.
However, Stokes also highlights two limitations of such AI-aided approaches. The first is that ‘many AI-generated structures remain impossible to synthesise’, which can result in expensive and time-consuming false starts. Secondly, the mechanism of action of AI-designed antibiotics needs to be unraveled, another technically challenging and expensive process. ‘But I’m certain the field will get to where it needs to go,’ he adds.
References
A Krishnan et al, Cell, 2025, DOI: 10.1016/j.cell.2025.07.033








No comments yet