Two of the smallest bacterial genomes ever sequenced.
Two of the smallest bacterial genomes sequenced to date provide new clues to the minimum number of genes required to support cellular life, report Japanese, American, and Spanish researchers.
In both cases, researchers sequenced the genomes of bacterial endosymbionts, microbes that live inside specialized cells of an animal host and provide that host with essential nutrients. Genome reduction is a common genetic feature of endosymbionts and the previous record holder for the smallest genome ever sequenced was about 600-kilobases in size and from a strain of Buchnera aphidicola, an endosymbiont of aphids.
Now scientists report the sequencing of a 160-kilobase genome from a strain of Carsonella rudii, an endosymbiont of psyllids, insects that feed on plant sap.1
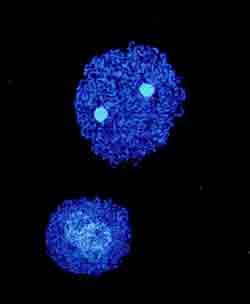
’In the case of C. rudii, the genome has gone beyond what seems possible for life,’ said Nancy Moran, an evolutionary biologist at the University of Arizona, Tucson. ’The genes that are missing are considered essential for basic tasks like replication, transcription, and translation,’ she explained. ’It’s still kind of a mystery how this bacterium survives.’
In addition to harboring only 182 genes, the arrangement of the genes is odd. ’The genes are very closely packed and most overlap by an average of 10 bases,’ explained Moran. ’This is another first for bacteria.’ Overlapping genes, where a few bases at a given location form part of more than one gene, are often found in genomes whose size is highly constrained (though they are also found, to a lesser degree, in higher organisms, including mammals). The vast majority, 90 per cent, of Carsonella genes overlap.
In a second study, scientists sequenced the 422-kilobase genome of a strain of B. aphidicola.2 Like the C. rudii genome, this genome has lost many essential genes and, surprisingly, the genes required for the synthesis of tryptophan, which is an essential amino acid that the endosymbiont produces for its host. A secondary symbiont of the aphid synthesizes the tryptophan needed by the host suggesting that B. aphidicola may be headed for extinction, explained Amparo Latorre, a geneticist at the University of Val?ncia, Spain.
’It is clear that B. aphidicola lacks genes that are essential for the host and that are now provided by the secondary symbiont,’ he explained. ’This is why we think B. aphidicola is losing its symbiotic capacity.’
Much remains to be learned about what drives the loss of genes from endosymbionts and how far the process can go before a cell is no longer functional. Studying tiny genomes can also be ’just plain interesting,’ said William Martin, an evolutionary biologist at Heinrich Heine University, D?sseldorf, Germany. ’It’s like collecting stamps, coins, orchids, or rare wines,’ he added, ’one can find particularly beautiful exemplars.’ Just like these two tiny genomes.
Jessica Ebert
Comparative genome analysis has changed theories about evolution and disease. Andrew West examines how it may also change ideas about drug development.
Double Nobel prize-winner Fred Sanger recalls his days in the lab, attributing his success to the time he spent at the bench. Emma Davies reports
References
1et al,Science3142et al, Science314, 312





No comments yet