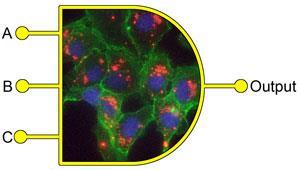
Synthetic biologists have already modified the genetic code of cells to create biocircuits capable of performing specific Boolean logic functions, for example AND gates and OR gates, for sensing, diagnostics and therapeutics. These genetic logic gates require sophisticated and extensive modifications of the cell DNA. Now, Erwin Märtlbauer and his team at the University of Munich have developed a comparably simple approach where proteins interact with the membrane of whole cells to produce a variety of combinatorial and sequential logic operators.
A unique enterotoxin protein made up of three components that must individually bind in a specific order to the cell membrane to cause cell death is central to Märtlbauer’s system. By using this sequential binding as the input of the logic gate and cell death as the output the team have created a logic operator with memory that is similar to a keypad lock, where unless the right key is pressed in the right order nothing will happen.
This strategy could be used to effectively steer cellular uptake or cell behaviour to construct more devices for cellular computation, but could also serve in diagnostic and therapeutic tools. ‘There are many bacterial toxins and they can be used as a toolkit to manipulate cells,’ says Märtlbauer. In principle it will be possible to determine the virulence of bacteria and even to selectively target and kill cells.
Lingchong You, a gene circuits and biocomputing expert at the Duke Institute for Genome Sciences & Policy in the US, is very interested in Märtlbauer’s approach. ‘It is an elegant example where the creative use of a limited number of biological components can lead to predictable programing of complex circuit functions.’
Substituting our standard desktop computer with biological computers is still a faraway dream, but this innovation will help push the limits of synthetic biology. The challenge now is to fully understand the basis of the protein ad cell interaction.




No comments yet