The threat of pandemic influenza is constantly on the horizon. Clare Sansom explores the latest attempts to tackle an ever-changing foe
The threat of pandemic influenza is constantly on the horizon. Clare Sansom explores the latest attempts to tackle an ever-changing foe
’I had a little bird,
Its name was Enza,
I opened up the window,
And In Flu Enza
This was a very common playground rhyme 90 or so years ago. It is not often that an outbreak of infectious disease becomes so deeply embedded in popular culture, but this was no ordinary outbreak. Between early 1918 and late 1919, pandemic influenza or ’Spanish flu’ killed forty million people worldwide: almost twice as many victims as the first world war.
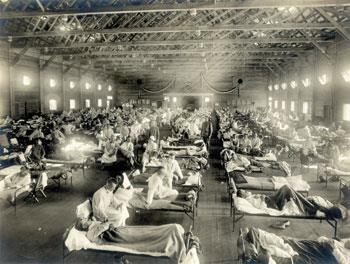
Influenza has a smaller place in the public imagination in our generation. Any public complacency, however, is not shared by the health community. Physicians have long memories; influenza pandemics killing hundreds of thousands worldwide occurred in 1957 and 1968, and continue to threaten today. In 2008, the UK National Risk Register described an influenza pandemic as ’the gravest threat to UK security [greater than terrorist threats or weather-related emergencies], as it could claim up to 750,000 lives’. This is despite the wide uptake of both vaccines and drugs. Both types of interventions have proved fairly successful, yet all known drugs and vaccines are effective only against a limited range of influenza viruses, and so far, at least, drug treatments have been limited by resistance.
The Ancient Greek physician Hippocrates was the first to describe the symptoms of influenza, in the fourth century BC. The first attempts to discern its cause, in the nineteenth century, fixed on a bacterium. The microbe is known - to the confusion of generations of students - as Haemophilus influenza but is unrelated to the disease. Influenza viruses were first discovered in the 1930s. They are roughly spherical viruses with RNA genomes bearing the characteristic surface proteins haemagglutinin and neuraminidase. These proteins recognise and bind to mono- and polysaccharide sugars on the surface of their target cells. They act at opposite ends of the viral life cycle. Haemagglutinin binding to cell surface sialic acid leads to the virus adhering to the cell membrane and then being engulfed by the cell, whereas neuraminidase catalyses cleavage of sialic acid from newly formed virus particles, releasing them from the cell. The drugs that are currently widely prescribed for influenza - zanamivir (GSK’s Relenza) and oseltamivir (marketed by Roche as Tamiflu) - are both neuraminidase inhibitors.
Mix and match
Another characteristic of influenza viruses is variability, and it is this feature that makes it so difficult to design universally effective drugs and vaccines. The classification of these viruses is complicated. The broadest classification is into three types: influenza A, B and C. Almost all the threat to human health comes from influenza A, which is also the only type to infect birds as well as mammals. Influenza A is further classified according to the variants of haemagglutinin and neuraminidase expressed on their surfaces, known as serotypes. So far, sixteen serotypes of haemagglutinin (named H1 to H16) and nine of neuraminidase (named N1 to N9) have been identified. These differ in the sugars that they bind to, the range of their bird and mammal hosts, and the clinical course of any infection. All combinations of H and N are found in birds, but only some infect humans and other mammals.
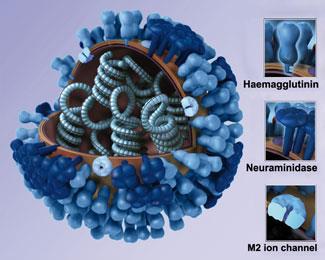
Variations in influenza A viruses arise through two mechanisms. Antigenic drift is the simple accumulation of mutations in the viral DNA that arise either at random or in response to drugs or the host immune response. However, if more than one type of virus is present in the same host population (most often birds or pigs), genes from the different viruses can cross over or ’reassort’ to form new types: this is known as antigenic shift. This may create a viral type that few humans have immunity to, and it is the main cause of human pandemics. So far, the only viruses known to cause major epidemics in humans contain haemagglutinin variants H1, H2, H3 and H5. The ’Spanish flu’ pandemic of 1918-19 coincided with the emergence of a new H1N1 strain; the 1957 pandemic was caused by a new H2N2 strain and the 1968 one by H3N2, which is still the most common viral subtype in circulation worldwide.
The next pandemic
The first decade of this century has seen not one but two feared influenza variants emerge, and this has spurred research into all aspects of the disease. The first is the H5N1 avian influenza (colloquially dubbed ’bird flu’) and the second a new H1N1 variant that arose in herds of pigs in Mexico, immediately becoming known as ’swine flu’. One remarkable feature of these two strains is how different they are in origin, pathogenesis and clinical course. H5N1 avian flu has infected only a few hundred humans since it was first characterised in 1997 in a small Hong Kong boy, and no cases of human to human transmission have yet been observed. It is, however, extremely pathogenic, with a fatality rate close to 50 per cent. In contrast, the H1N1 variant that arose in Mexico in 2009 is exceptionally infectious but generally causes mild symptoms.

’The greatly feared word "pandemic" is used to mean any infection that has spread worldwide’, says John McCauley, director of the World Health Organization (WHO) Influenza Centre at the National Institute for Medical Research in London, UK. ’It was correct to call the 2009 epidemic a pandemic, even though few were badly affected.’ The few people who did contract severe illness, however, were young or middle-aged. This pattern, which is opposite to that observed in seasonal flu epidemics, derives from the virus’s H1N1 serotype. ’H1N1 was the main virus in circulation before 1957’, says McCaulay. ’Almost everyone who is now over 60 will have been exposed to this serotype before and will therefore have acquired some immunity.’
Of these emerging influenza variants, therefore, one is highly pathogenic but not very transmissible; the other is highly infectious, but causes mild disease. Any reassortment between genes from these viruses to produce a subtype with the infectivity of H1N1 and the pathogenicity of H5N1 would clearly cause a public health nightmare. Perhaps reassuringly, Ilaria Capua from the World Organisation for Animal Health and United Nations Food and Agriculture Organization Reference Laboratory for Avian Influenza in Padova, Italy, recently claimed in a keynote conference lecture that ’Despite its very wide host range, the H5N1 virus doesn’t readily reassort with other subtypes’: nevertheless, this cannot be completely ruled out.
Prevention is better than cure
Vaccination has been the mainstay of influenza treatment for many decades. The technology used for vaccine production, however, has scarcely changed since the 1950s: the vaccine produced for each ’flu season’ consists of the two strains of influenza A (usually, one H1N1 and one H3N2) and one of influenza B predicted to be most highly circulating, grown in fertilised chicken eggs. This is a slow process, so strain predictions need to be made over six months before the vaccine is needed. Until - or unless - the much heralded ’universal influenza vaccine’ ever becomes available, researchers are studying to improve the technology to shorten this lead time, and using bioinformatics to improve predictions. A vaccine strain produced by US pharmaceutical firm Baxter in African green monkey (Vero) cells has recently been licensed.
Vaccines that generate antibodies against more conserved regions of influenza viruses should also be more widely effective. ’Generating antibodies that target the haemagglutinin ecto-domain, which is conserved in many different strains, may be seen as a "conceptual advance",’ says Vladimir Brusic of the Dana-Farber Cancer Institute, in Boston, US.
As well as vaccines, companies have looked to small molecule drugs to suppress the viruses in infected patients. The first anti-influenza drug, amantadine, was licensed in the 1960s. This targets the influenza protein M2, a channel in the virus membrane that allows protons to enter the cell; interestingly, it is also an effective drug for Parkinson’s disease. A very similar molecule, rimantadine, was licensed to treat influenza in 1994. Influenza viruses rapidly become resistant to these drugs, however, and they are now very rarely prescribed.
During the 1970s and early 1980s, the UK drug companies Glaxo and Wellcome, now both part of the giant GSK, were at the forefront of antiviral drug research. There seemed, however, to be few reasons to focus on influenza. Dick Challand, a medicinal chemist working at Wellcome at that time, remembers that the disease was thought of as ’a ’white elephant - a largely self-limiting infection that could be well controlled by vaccination’. Nevertheless, Wellcome took an inhibitor of viral RNA polymerase into pre-clinical development.
Interest switched to the viral surface proteins when their three-dimensional structures became available later in the 1980s. The neuraminidase inhibitor zanamivir was discovered by a group led by Mark von Itzstein at Monash University, Australia, using structure-based drug design and developed by the Australian biotech company Biota and Glaxo (later GlaxoWellcome) in the UK; it was first marketed in 1999, a year before GSK was formed by the merger of GlaxoWellcome with SmithKline Beecham. ’Glaxo had only a few months’ market lead with zanamivir over Roche’s rival compound, oseltamivir’, explains Challand. ’Oseltamivir is much more prone to developing resistance, but it is currently the market leader because it is orally available [whereas zanamivir must be inhaled].’
Certainly when, in 2009, many governments stockpiled large quantities of drugs in advance of the predicted pandemic, oseltamivir was by far the more popular choice. Several papers published since the pandemic have documented increasing resistance to it. A third neuraminidase inhibitor, peramivir, is now being developed by BioCryst Pharmaceuticals in Birmingham, Alabama, US.
Widening the net
With resistance to neuraminidase inhibitors being reported more frequently, influenza researchers have begun to explore new targets. They are also re-visiting the M2 proton channel, helped again by new insights into this protein’s structure. Several independent groups have determined atomic resolution structures of M2, but the results of these have often proved inconclusive, particularly in relation to drug binding. Writing recently in a commentary in Science, Bill DeGrado of the University of Pennsylvania, US, claimed that the debate was ’settling in favour of a pharmacologically relevant [amantadine binding] site in the M2 channel pore’, but that there was still controversy about, among other things, the protein’s mechanism of action.
Mei Hong and her team at Iowa State University, US, obtained the M2 structures that resolved this part of the controversy. ’We know that in most currently circulating influenza viruses, one serine residue has been mutated to asparagine, and that this mutation confers resistance to amantadine’, she says. ’We were very pleased to see amantadine bound to this serine in our wild type structure.’ A lower affinity binding mode with the drug outside the channel has also been observed. Hong’s NMR methods can be used to visualise drug-protein binding at high resolution in an environment that mimics the viral membrane. ’We can observe subtle structural differences between drug binding modes that may help predict the potential success of new antiviral compounds’, she says. However, none of the compounds that have so far been tested have been found to be potent and selective inhibitors of amantadine-resistant M2.
Superficially, at least, haemagglutinin would seem to be a good drug target, in some ways better than neuraminidase, as it acts at the beginning rather than the end of the viral life cycle. The main difficulty with designing drugs to inhibit haemagglutinin binding is that the haemagglutinin receptor binding domain (RBD), which is the obvious target of such molecules, differs widely between serotypes. ’We cannot expect a drug designed to target the haemagglutinin RBD of one influenza strain to be active against other strains’, says Dong Xu, assistant professor of chemistry at Boise State University, Idaho, US. Using molecular dynamics simulations, Xu has discovered that all RBDs bind to a common ’fish-hook like’ shape on the receptor surface. ’We are now searching for compounds that mimic this glycan receptor shape’, he says. His group is also investigating the viability of designing molecules to target other sites on haemagglutinin.
So far, it would be safe to say of both recent influenza outbreaks, H5N1 ’avian’ and H1N1 ’swine’ influenzas, that their bark has been worse than their bite - at least at population level. Yet pandemics of highly pathogenic influenza remain a serious threat. If the increased media prominence of influenza has inspired any of the innovative work in drug and vaccine discovery currently underway, that is all to the good. Another positive legacy of the avian flu scare has recently been highlighted by Capua: increased cooperation between researchers into human and veterinary infectious disease.
Clare Sansom is a freelance science writer based in Cambridge, UK






No comments yet