Answers to how living cells and bacteria receive molecular signals and deal with the initial steps of viral infection may soon be revealed.
Answers to how living cells and bacteria receive molecular signals and deal with the initial steps of viral infection may soon be revealed. Scientists at the University of Twente, the Netherlands, investigated complex multivalent interactions using cyclodextrin as a receptor molecule. They hoped to shed light on molecular recognition as it occurs in nature.
Bart Ravoo and colleagues showed that small messenger molecules bind through a number of weak interactions to cyclodextrin receptor molecules on the surface of an artificial membrane. Together these interactions create a very strong bond. The small molecules also appeared to bind selectively to specific receptors, allowing them to bind much more efficiently than expected.
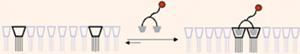
In nature, this type of messenger molecule transfers information between cells and compartments. A virus that infects a cell or bacteria also uses the same principle to bind to the cell’s surface. This new cyclodextrin model therefore paves the way for better understanding of molecular recognition in nature.
This highly selective molecular recognition may be used by scientists in the future to bind not only small molecules to membrane vesicles, but also one type of membrane vesicle to another. It may even be possible to fabricate arrays of nanosized containers entirely by self-assembly from all the necessary components.
But nature is a complicated process and matching the specificity and selectivity in molecular recognition at the cell membrane is still a big challenge. Ravoo foresees the major test as being able to overcome the dynamic nature of the system. ’All components tend to move.with this making it difficult to use them to make a structure with a well-defined shape and long-term stability,’ he said.
Jenna Wilson
References
C W Lim, B J Ravoo and D N Reinhoudt, Chem. Commun., 2005, (DOI: 10.1039/<MAN>b510540d</MAN>)






No comments yet