A new technique for the accurate and fast detection of amino acids has been developed by researchers in the US. The simple method, employing cucurbiturils and the lanthanide europium, could light the way to improvements in nutritional analysis and drug detection.
Amino acids are essential building blocks for all living beings. Abnormal amino acid levels can indicate underlying health problems such as malnutrition, pancreatitis and Alzheimer’s disease. Conventional amino acid sensing techniques involve chromatography or electrochemical methods, which can be costly and must be performed by skilled operators.
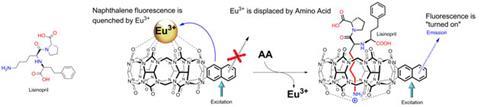
The method proposed by Pavel Anzenbacher Jr and coworkers at Bowling Green State University in Ohio analyses fluorescence signals when guest europium ions are displaced from an array of two cucurbituril host probes. One of the probes is cylindrical and prefers to bind smaller molecules in a host–guest fashion, whilst the other is a larger linear molecule that wraps around larger analytes.
The europium ion quenches the fluorescence of the probes, leaving the sensor in the “off” state by default. When an amino acid binds to the probe, the europium is displaced and the sensor is “turned-on”, allowing the probe’s natural fluorescence to be visible again.
Different analytes bind to the two probes with different affinities so the degree of fluorescence is unique to the guest. Statistical analysis of the fluorescence signals correctly identified 10 analytes including amino acids, their corresponding amines, and the tri-amino acid drug molecule lisinopril, with a 100% success rate.
‘In future we are looking at using this type of system to detect different side chains on proteins,’ says Anzenbacher. ‘With four probes for example, we could detect 50–100 different analytes.’ He goes on to add that ‘the real strength of this analysis is that we are able to differentiate between groups of analytes, such as amino acids and their corresponding amines.’
“Turn-on” sensors have the advantage of being more specific than the more passive “turn-off” methods. While the “turn-off” sensors work by starting off in a fluorescent state that can be quenched on the binding of any unknown compound in the mixture, “turn-on” sensors are only activated when the correct analyte binds.
Ramon Vilar, an expert in biomedical applications of supramolecular assemblies, including molecular probes for biosensing, at Imperial College London in the UK, comments that this is an exciting way forward for amino acid detection. ‘However’, he cautions, ‘this type of sensing relies on the ‘fingerprint’ signal of the molecules being detected, and as such, their detection in complex mixtures such as cell lysate or blood samples is not trivial.’ Anzenbacher agrees that this will be tricky, but highlights that work is being undertaken in his lab to develop “turn-on” probes for applications such as drug detection in urine samples.






No comments yet