Simulations suggest that electrostatic and non-electrostatic forces contribute equally to make DNA one of nature's most rigid polymers
Computational chemists believe they have solved one of the puzzles about the stiffness of DNA: how much of the molecule’s rigidity is due to electrostatic forces and how much to its physical structure.
Double-stranded DNA is one of nature’s stiffest polymers. The flexibility of the molecule is of interest to biologists because the extremely long structure must be compacted and folded so that it can fit into a relatively tiny space within the nucleus.
’The compaction of DNA is important in areas such as virology, where the molecule must fit into the virus shell, and gene therapy, where we need to find ways to package and deliver DNA,’ says Garegin Papoian of the University of North Carolina in the US, who led the work.
There is wide agreement that two main factors account for the molecule’s rigidity. Mechanically the base pair building blocks of the polymer are stacked together with sufficient force to impart stiffness to the structure. Allied to this is the fact that DNA has negative charges studded down its length thanks to the molecule’s phosphate groups. These repel each other electrostatically, so that any bending of the molecule will bring the charges closer together - a situation that will be resisted. The relative contribution of each of these factors has, however, remained controversial.
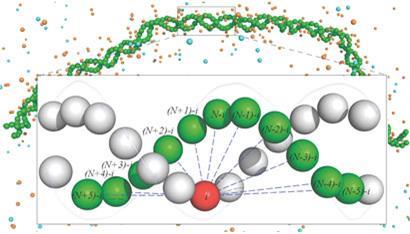
To resolve the issue the researchers constructed a high resolution atomistic simulation of DNA. ’We represent every atom in the molecule and the surrounding environment and run a molecular dynamics simulation to see how it wriggles around,’ says Papoian. ’We can then simulate protonation of the phosphates, making them neutral, and see what happens.’ In this way the neutral isomer was seen to be more flexible than its charged counterpart. The experiment was repeated with a coarser-grained model of DNA, which the team had previously shown could still simulate the important features of the molecule. In this case a gradual neutralisation could be simulated, with an increasing degree of flexibility observed.
It appears from these computational studies that the contribution of the electrostatic and non-electrostatic forces to DNA’s rigidity are more or less equal, says Papoian.
Sarah Harris, who studies the biophysics of DNA at the University of Leeds in the UK, says that the finding could have implications for approaches to manipulating DNA. ’Electrostatic interactions can be easily modified by changes in the environment, and can be reduced by increasing the salt concentration, or by interactions with other molecules, such as charged DNA binding proteins, which would serve to decrease the rigidity of the molecule making it easier to pack,’ she says. ’It is not so easy to see how this can be achieved if the base-stacking interactions also play a significant role.’
Simon Hadlington
References
DOI, 10.1021/ja207984z






No comments yet