Understanding how bacteria communicate has particular relevance to understanding how pathogenic types cause disease progression. Now, Jason Shear and colleagues at the University of Texas, US, have developed a 3D printing technique that lets them ‘construct’ defined bacterial communities so that short-range chemical communications and physical interactions between bacteria can be investigated more systematically than ever before.
Most laboratory studies of bacterial communications involve monitoring chemical signals, oxygen demands and other factors averaged over a large population. But specific cell-to-cell interactions could provide new insights into the way in which individual bacterial cells respond to their neighbours, as well as to external factors. This is particularly important given that pathogenic bacteria often exist in three-dimensional structured colonies of multiple species in our bodies. The 3D printing technique allows the team to organise multiple populations of bacteria in almost any geometry for their experiments.
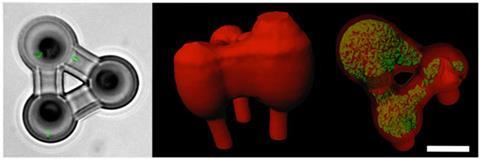
The team explains that they can use laser-based lithographic techniques with microscale resolution to carry out cross-linking of highly porous gelatin in which selected bacteria are suspended in tiny containers. The cross-linking of polypeptides within the gel locks the bacteria into position so that they can create adjacent, nested and free-floating colonies. The highly porous nature of the gelatin matrix allows the bacteria to multiply freely from the central fixed cell in each cavity within the gel. The researchers point out that this porosity also allows free movement of signalling molecules between cells and the diffusion of added chemicals, such as antibiotics.
The approach opens up the possibility of developing new experimental setups in which such 3D printed bacterial colonies are created within microfluidic devices for specific tests and assays. As a proof-of-principle, the team has demonstrated how they can create picolitre volume aggregates of the bacterium Staphylococcus aureus enclosed within a shell of a second bacterium, Pseudomonas aeruginosa. They can then easily infuse this system with beta-lactam antibiotics. They found that the presence of the second microbe strongly enhances the resistance of S. aureus to the antibiotics. Such findings could change how doctors treat multiple bacterial infections in people with cystic fibrosis and other conditions, the team suggests.
‘There is a rapidly growing community focused on “biofabricating” the assembly of biological components into structures that will transform our ability to understand signalling phenomena at nature's length and time scales,’ says bioengineer William Bentley of the University of Maryland, US. ‘This is exciting work that opens new avenues of exploration that have not existed and hold great promise to provide a platform for new discovery. The problems of antibiotic resistance are huge and the right tools for study are few.’





No comments yet