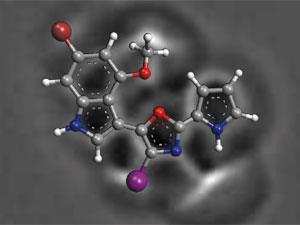
A combination of atomic force microscopy (AFM) and computational techniques have been used to solve the unusual structures of two natural products from the Arctic coral-like animal, Thuiaria breitfussi. The compounds, known as breitfussins, have been difficult to study as they are only available in small amounts, preventing analysis by x-ray crystallography. Previously, researchers have used the standard techniques of mass spectrometry and NMR to identify indole, oxazole and pyrrole aromatic groups but could not piece them together.
By imaging the breitfussin molecules using AFM, the connections between the groups were clear and revealed that their unusual structures could be derived from the dipeptide proline–tryptophan. This technique was employed by the same team from IBM who previously rose to fame with their 2009 paper in Science where they were able to image molecules with individual atoms resolved. However, AFM was unable to differentiate between the oxygen and nitrogen in the oxazole ring and so the researchers could not place them confidently. Using computational approaches, the researchers calculated the expected 1H and 13C NMR chemical shifts and compared them to the experimental values to determine their positions.






No comments yet