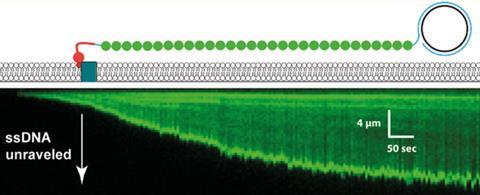
When one end of double-stranded DNA is attached to the surface of the device and a flow is applied, the DNA stretches out into a line. With enough strands, the DNA forms a ‘curtain’ hanging from a rail. This allows scientists to study the dynamics of single protein molecules interacting with the DNA in real time.
However, single-stranded DNA is flexible and tends to form hydrogen bonds with itself, resulting in a disorganised length of loops, twists and turns (those readers with unmanageable hair may empathise). This is why single-stranded DNA has been so unsuitable for forming curtains previously.
Researchers from Columbia University avoided this problem by employing a fluorescently-tagged version of replication protein A (RPA). This protein binds tightly and specifically to ssDNA, causing it to lose its tangled structure and become less flexible, making it suitable for forming a ‘curtain’ – think Frizz Ease for DNA. By fluorescently labelling the protein, it also made observing the ssDNA simpler.
Eric Greene, who led the project, explained the motivation for developing this technique. ‘For years we’ve been studying the proteins involved in homologous recombination using dsDNA as the substrate and every time I talked about this, the number one question was: “Can you do the same experiments with ssDNA?”’
As a proof of principle, a helicase protein, Sgs1, was tethered to a magenta-coloured quantum dot to do a two-colour experiment. The researchers were able to show that the labelled Sgs1 and RPA proteins bound along the whole 50,000 nucleotide lengths of ssDNA. Greene is already looking to extend the capability beyond this, ‘we’ll probably move towards three colour imaging in the coming couple years’.
Malcolm White from the University of St Andrews, UK, who was not involved in the research, commented: ‘It’s really interesting to know that it’s possible now to look at single-stranded DNA in the same way that double-stranded DNA has been visualised for maybe 10 years... in that respect it’s a significant advance.’ He suggested that this new technique would allow researchers to further their understanding of DNA repair and recombination. ‘By using a variety of fluorescently labelled proteins and putting them in complexes, we can see the choreography of these very complex pathways for repair, for example, in an unprecedented level of detail.’




No comments yet