Joint industrial–academic team develops and validates force-field tool for identifying ligands for enantioselective catalysis
Chemists in the UK, Sweden and the US have shown that their easy-to-use tool can quickly predict which catalysts will selectively produce one of two possible enantiomer forms of molecules. The tool, CatVS, virtually screens substrate and ligand libraries within hours, says Olaf Wiest from the University of Notre Dame, US. ‘All they need to do is plug the molecule into a website,’ explains Wiest. ‘It’s accurate and it’s predictive and it’s fast.’
Wiest and his colleagues at AstraZeneca’s Macclesfield, UK, and Mölndal, Sweden, sites constructed CatVS by building on transition state force field-based virtual screening methods already used by chemists to design drugs. Such molecular mechanics approaches treat atoms as balls and the bonds between them as springs, modelling them using classical Newtonian physics to keep the calculations involved manageable.
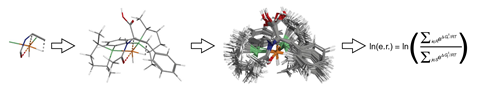
But in avoiding dealing with quantum properties that approach prevents scientists studying the transition states at the heart of chemical reactivity and catalysis. ‘This thing doesn’t know what an electron is,’ Wiest says. ‘So how do you treat a reaction where you have bond-making, bond-breaking?’
The answer is quantum guided molecular mechanics (Q2MM), developed by AstraZeneca’s Per-Ola Norrby. Q2MM starts by using more time-consuming density functional theory (DFT) quantum mechanical calculations to define transition state force fields suitable for virtual screening. CatVS enables non-expert chemists to integrate force fields with substrates and ligands and quickly do “conformational sampling”.
CatVS screened 48 combinations for an osmium tetroxide cis-dihydroxylation catalyst generated by four stereoisomers of a ligand and 12 substrates in around seven hours, with results matching earlier experiments. The researchers also virtually screened ligands and substrates for rhodium-catalysed asymmetric enamide hydrogenation. Experimental work, led by Rachel Munday in the AstraZeneca Catalysis Hub, then showed that four of five ligands CatVS predicted to offer enantiomeric ratios exceeding 98:2 actually did so.
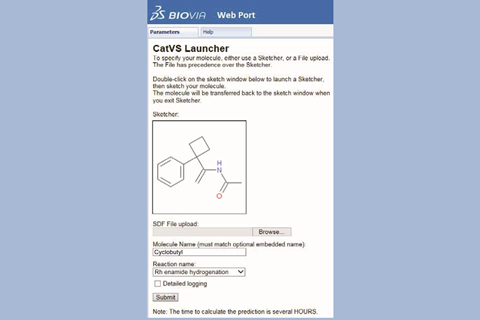
Producing the initial force fields using DFT is still a bottleneck, however, with the team having done around a dozen so far. The researchers are therefore trying to simplify that, while Wiest also wants to hear from potential CatVS users about which catalyst systems they’d like force fields for. ‘What reactions are you interested in?’ he asks.
Identifying stereoselective catalysts through computational screening will have obvious utility, says computational chemist Steven Wheeler from the University of Georgia, US. ‘However, the approach would be even more powerful if it could be used to predict catalytic activity in addition to selectivity,’ he adds.
References
A R Rosales at al,Nat. Cat., 2018, DOI: 10.1038/s41929-018-0193-3














No comments yet