Scientists use mirror-image protein to identify potential new drugs from a previously unexplored library of natural product enantiomers
Researchers have exploited the structural symmetry of natural products and proteins to access a vast virtual library of mirror-image compounds in a smart new approach to drug discovery.1
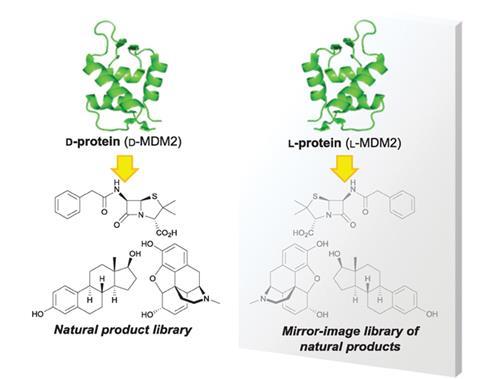
Scientists screen huge chemical libraries in search of compounds that interact with a given protein and have the potential to become clinical drugs. However, as all library compounds need to be individually synthesised or isolated from natural sources, researchers often only test a single enantiomer of chiral compounds. In their new approach, Shinya Oishi and colleagues at Kyoto University and RIKEN, Japan, examined readily available natural products to interact with an unnatural, mirror-image protein. Once they found a lead compound, the team synthesised the mirror-image structure and evaluated its fit with the natural protein, because the two proteins fold in exactly opposite ways.
Using standard peptide coupling reagents,2 Oishi’s team synthesised the mirror-image (d-MDM2) of natural protein MDM2, which regulates a tumour suppressant in the body. They then screened the unnatural protein against a library of 22,293 compounds from the RIKEN Natural Products Depository. Having identified four compounds that inhibit d-MDM2, Oishi’s team synthesised the corresponding mirror-image enantiomers, which indeed blocked natural MDM2.
For Oishi, his method’s key advantage is that it minimises the time consuming and costly synthesis during drug discovery. ‘No one has had facile access to the mirror-image library of chiral small molecules. Using our approach, we can evaluate this library without the synthesis of large numbers of mirror-image compounds,’ Oishi says. Intriguingly, the approach could be extended to targets such as enzymes and receptors once scientists develop a straightforward way to make the mirror-image biomolecules.
Brian Cox, who carries out research in chemical synthesis and drug discovery at the University of Sussex, UK, describes the study as ‘a very exciting approach, assuming technologies can provide the proteins for screening’. However, he notes that there is no guarantee the virtual library will deliver a rich source of new lead compounds.
Producing sufficient quantities of the mirror-image compounds needed for evaluating efficacy and safety in humans is a potential drawback, says Greg Challis, whose research at the University of Warwick focuses on natural product chemistry and biology. On the other hand, he suggests ‘it may be possible to use the information obtained to design simplified analogues of the unnatural enantiomer that are easier to synthesise, yet retain good levels of bioactivity’.
References
1 T Noguchi et al, Chem. Commun., 2016, DOI: 10.1039/c6cc03114e This article is open access
2 T Noguchi et al, Bioorg. Med. Chem. Lett., 2013, 23, 3802 (DOI: 10.1016/j.bmcl.2013.04.094)









No comments yet