
X-ray crystallography has shaped modern chemistry. It is arguably the most powerful tool for molecular structural analysis. But it suffers from one big drawback: it can only analyse materials that form well-defined crystals. This may now be about to change. Researchers in Japan have used ‘crystal sponges’ to hold molecules that can’t be crystallised, allowing them to be analysed using x-ray crystallography.
For chemists, trying and failing to crystallise compounds can be a frustrating task. Yasuhide Inokuma’s experience was no different. So, in 2010, Inokuma and colleagues at the University of Tokyo came up with a solution. With the help of metal atoms and organic ligands they built molecular cages, more commonly known as metal-organic framework (MOF), to trap fullerenes and analyse their structure using single crystal diffraction (SCD), a type of x-ray crystallography.
Now, they have gone a step further and tuned the molecular cages to enable SCD of all sorts of compounds, even those that are liquid at room temperature. Remarkably, this can be done with less than 0.1µg of the compound.
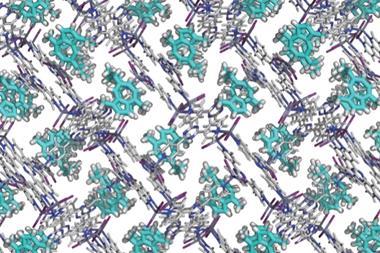
Inokuma and colleagues built the molecular cage using either cobalt(II) thiocyanate or zinc(II) iodide with tris(4-pyridyl)- 1,3,5-triazine to form an octahedral cage. The target molecule can be caged by dipping a single crystal of the MOF into a solution of it. Once inside the MOF, the target molecule settles into the most thermodynamically stable orientation. This is key, because SCD only works when molecules are held in the same orientation to allow the x-rays to scatter in a reliable manner.
Parsimony for stereochemistry
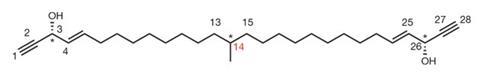
In their initial attempts they were able to successfully trap cyclohexanone and isoprene, both liquids at room temperature, and analyse their structures. Next they took up the challenge of analysing the structure of miyakosyne A, a long chain marine natural product. With only 5µg to hand, they were not only able to nail down its structure, but also determined the absolute stereochemistry of its hydroxy groups.
There are limitations, however. Inokuma admits that ‘data from pure single crystals, that is without using molecular cages, are usually better than our method, which is to say that bond length and angle are more reliable’. But for the many compounds that do not crystallise at all, this method is still good enough to obtain their structure, especially when used in conjunction with standard tools like NMR and mass spectrometry.
In recent years, combining separation with analysis – LC–MS, for instance – has proven invaluable for chemists. Inokuma and colleagues envisage LC–SCD, combining liquid chromatography with single crystal diffraction to create a powerful new analytical tool.
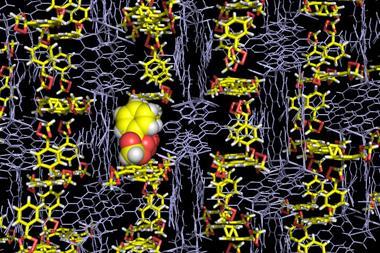
They’ve already trialled their LC–SCD technique on dried orange peel. The peel extract was run through an HPLC machine, with each compound delivered into a vial containing a single crystal of the molecular cage. The team were then able to get good structural data on the peel molecules.
The revolution will not be crystallised
So, could Inokuma’s method revolutionise x-ray crystallography? ‘It certainly could,’ says Richard Cooper, head of chemical crystallography at the University of Oxford, UK. ‘There will, as ever, be caveats and exceptions, but this certainly pushes the envelope of what can be studied by SCD in the right direction.’
But Amber Thompson, another crystallographer at Oxford, is less enthused. ‘The key is matching the size and surface properties of the cavity with the target molecule,’ she says. Thus, if the cavity is too big the target molecule will ‘flop’ about rendering the analysis useless. If it is too small the target molecule may not fit in the cavity.
Inokuma admits that very large molecules, such as proteins, can’t currently be analysed. ‘We have to work to make our method more versatile,’ he says. ‘But we hope that in five years our method will be standard protocol.’




No comments yet