Protein cage structure shows the way to building biomolecular architectures
Researchers in the US have designed a hollow cube out of naturally occurring proteins, something that was previously only possible with DNA. The custom-built protein structures might be used for drug delivery, or as reaction or crystallisation vessels.
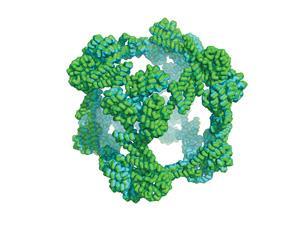
Researchers have been engineering DNA into custom nanostructures (DNA origami) for several years, but proteins have proved more challenging. ‘We don’t yet fully understand the rules that govern how proteins fold and bind to each other. DNA structures have surged ahead in the last decade because their rules of assembly are a lot simpler,’ explains Todd Yeates from the University of California Los Angeles, US. Previous approaches to ‘protein origami’ relied on designing new, unnatural protein oligomers, a challenge Yeates and his colleagues sought to avoid.
Instead, Yeates and his team exploited proteins that naturally assemble into oligomers. They fused together two different proteins, one which forms a dimer and the other a trimer, using a short amino acid linker. ‘If you get the geometry just right, the [fused] protein won’t have any choice but to assemble into the desired shape,’ Yeates explains.
Having used computational analysis to identify which proteins and amino acid linkers would satisfy their design criteria, the researchers genetically modified Escherichia coli bacteria to express the desired protein sequence. After purification, the fused proteins assembled into a hollow cage, with the trimers occupying the cube’s corners and the dimers forming the edges. This design is the latest advance in efforts by multiple laboratories to create large geometric protein structures.
The protein cages are a conceptual novelty, but Yeates thinks they could have applications in synthetic vaccines or drug delivery. ‘We could put things inside these structures or attach things to the outside,’ he says, ‘imagination is the limit.’ Scientists have used other highly porous materials, like metal-organic frameworks (MOFs), for gas storage or as sensors, but their biological compatibility is limited. The cavity inside Yeates’ protein cage is also unusually large compared to MOFs, measuring 130Å. ‘One of the reasons for thinking protein structures will be better than DNA structures or MOFs is that biology chose proteins as its workhorse,’ adds Yeates. Roman Jerala from the National Institute of Chemistry, Ljubljana, Slovenia, who designed a tetrahedral protein cage in 2013, says that proteins main advantage of over DNA is ‘their versatility – there are 20 amino acids with different functional groups compared with only four nucleic acids’.
Jerala agrees that there are many potential applications to explore. ‘Perhaps there will even be hybrid structures, combining proteins, DNA and other nanomaterials,’ he says. ‘There are countless possibilities.’











No comments yet