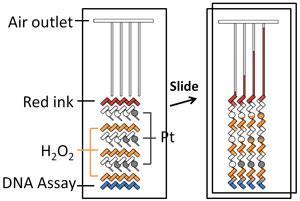
The system consists of a narrow, sealed channel around 80µm across etched into a glass slide and punctuated by a series of reservoirs and reaction chambers. A short strand of DNA complementary to the target sequence is immobilised in a reaction well at the start of the channel. The solution containing the target sequence is introduced, which binds to the immobilised strand. This traps the target DNA. A second strand of DNA, complementary to a different part of the target DNA, is then placed in the well. This strand has the enzyme catalase covalently bound to it. The effect is to bind catalase to the target DNA: the concentration of catalase present in the reaction well is therefore dictated by the amount of target DNA.
The chip is then manipulated to open up the channel system, allowing hydrogen peroxide to enter from an adjacent reservoir. The catalase reacts with the peroxide to release oxygen. This in turn pushes up the channel, meeting a second reservoir of hydrogen peroxide, which it forces into contact with a deposit of platinum. This reacts to produce yet more oxygen, pushing it upwards to meet another store of hydrogen peroxide, which in turn is propelled on to another island of platinum. This process is repeated a number of times, each time amplifying the initial signal produced in the first reaction well containing the DNA.
The final pulse of oxygen comes up against a reservoir of ink, which it forces upwards to provide a visible read-out: the more target DNA present, the more oxygen generated, the further up the channel the thread of ink travels. The system can be configured to run multiple samples simultaneously side-by-side by increasing the number of channels in parallel.
The ‘rocket-propulsion’ technique can amplify the signal more than a thousand times, says lead researcher Lidong Qin. The main advantages of the system, says Qin, are that it is portable, inexpensive, has a quantitative read-out and does not require sophisticated instrumentation.
For Rob O’Brien, of the National Forensic Science Technology Center in the US, the main drawback is the time it takes to process the sample. ‘The method proposed requires two separate incubation times of one hour each followed by some manual manipulation,’ he notes. ‘Current methods used in forensic quantitation of DNA take less than two hours to quantify up to 80 samples and there is technology reducing this time to 30 minutes or less. Therefore, even though this may be a promising method for quantitation of DNA in forensics, high throughput and faster quantitation times rule the day.’




No comments yet