DNA nanotechnology has moved a long way since its first public appearance in 1991 - and its first applications are already on the horizon, says Michael Gross
DNA nanotechnology has moved a long way since its first public appearance in 1991 - and its first applications are already on the horizon, says Michael Gross
Structural DNA nanotechnology - or the ’misuse of DNA’ - was arguably started as a research field when Ned Seeman used DNA strands to built a nanoscale cube in 1991.1 ’At the time, we had control only of the topology of objects, but not their structures,’ Seeman reminisces.

Seeman and his coworker Junghuei Chen could not provide enough material to solve the three-dimensional structure of the new species at the time, but what they did prove was that the methods of molecular biology, revolutionised in the 1980s, can be used for other purposes apart from genetics and cell biology. From then on, more and more scientists have begun using DNA as a molecular-scale building material.
Yet despite passing several more milestones - including DNA computation, demonstrated by Leonard Adleman in 1994,2 and the diagnostic-therapeutic ’DNA doctor’ developed by Ehud Shapiro’s group in 2004 to detect and respond to cancer biomarkers3 - DNA nanotechnology continues to present itself as a frontier where exploratory research dominates and applications are forever in a hazy future. Now that the field is turning 18, is it about to come of age?
Programming structure
The mainstay of what Seeman defines as ’structural DNA nanotechnology’ involves ’using DNA as the bricks and the mortar to build things,’ Seeman explains. However, the methods of construction have become more sophisticated over time. Seeman built his original cube and similar structures in the 1990s using ligation to clip together fragments of double stranded DNA - which makes the whole process slow and a bit tricky. In recent years, alternative approaches based on self-assembly, where two complementary single strands pair up to form a particular architecture without any changes in covalent bonds, have become very popular in the field.

One such approach is known as DNA origami and is based on an asymmetrical pairing. One long strand of DNA, the scaffold strand, forms the underlying structure, which is forced into the desired shape by adding other, shorter strands known as staple strands.
Paul Rothemund at CalTech in the US used this origami method to fold a strand of natural, genomic DNA (from the virus MP13mp18) into two-dimensional patterns visible in electron micrographs and AFM scans. In March 2006, his 2D DNA images, including stars, triangles, and smiley faces appeared on the cover of Nature.4 With dedicated computer algorithms, researchers can now design staple strands that turn any DNA of a given length into any two-dimensional object. Maps of the world and of particular countries have been popular targets among researchers trying out this approach.
Taking the origami to the third dimension, Chengde Mao and coworkers at Purdue University, US, produced flexible three-point stars with ’sticky ends’ - short strands of unpaired DNA. These unpaired ends can be used to join multiple stars together to build up three-dimensional objects including tetrahedra, dodecahedra, and buckyballs. The structure that preferentially forms can be controlled by adjusting the flexibility of the edges carrying the sticky ends and on the concentration of the units.5
Hao Yan at Arizona State University in the US has taken the origami technique a step further to create functioning molecular probes, which could become the nanoscale equivalent of DNA microarrays. Like Rothemund, Yan used a long single strand DNA as the scaffold and stapled it into the desired shape using short strands. To allow the construct to detect a target RNA, Yan created specific binding sites by adding the complementary sequence to the free ends of the short staple strands - but split the sequence across two adjacent staples. When the target RNA binds across the two ’half probes’, it is forced into a V-shaped conformation, which is stiff and readily detected by atomic force microscopy.
Yan’s group showed that their probe could selectively detect the target RNA sequence in a solution containing large amounts of cellular RNA, and also that the probe could be designed to incorporate several sets of probes, detecting multiple RNA targets.6
Making use of DNA dynamics
While DNA is the key building material used in the field, the increasing interest in DNA nanotechnology has also led to developments of hybrid structures where DNA is combined with other materials. One recent development uses DNA’s flexibility and dynamics as much as its sequence programmability.
Last year, two US-based teams simultaneously came up with such a dynamic DNA architecture. Both Oleg Gang at the Brookhaven National Laboratory in New York State and Chad Mirkin at Northwestern University in Evanston, Illinois, set out to arrange gold nanoparticles in crystal-like three-dimensional arrays using DNA linkers.7,8
Both groups hooked up one half of their nanoparticles with a given single stranded DNA sequence (A) and the other half with the complementary sequence (B). This arrangement ensured that A particles could bind to B particles, but neither would bind to their own kind.
Both teams found that their highly flexible DNA linkers allow the formation of highly ordered structures. For optimal ’crystallisation’, a flexible linker between the gold particle and the specific binding sequence turned out to be essential. The structure formation also benefited from gentle warming, favouring a dynamic equilibrium between dissolution and renewed binding of the sequences.
With this kind of encouragement, the gold particles equipped with flexible arms readily shook hands to line up and form familiar three-dimensional structures such as face-centred cubic or space-centred cubic arrays, depending on the precise conditions. Unlike the strictly designed DNA shapes produced since the early 1990s, this approach allows a random ’cloud’ of DNA strands to surround each gold nanoparticle, while still achieving highly regular ordered arrays of the particles thanks to equilibrium thermodynamics.
Programming function
A notable trend in recent years has been the construction of DNA machines, equipped with DNA ’hands’ to manipulate molecular objects, or ’feet’ to walk across DNA substrates.
Hermann Gaub’s group at the Ludwig Maximilian University at Munich, Germany, for instance, transformed an atomic force microscope into a DNA-based crane, which can pick up molecular building blocks from a reservoir and deposit them in a target area.
The researchers used a fluorescent dye to demonstrate the function of their crane. They fitted this shiny cargo with two DNA tools, one to hold it in place while stored at a ’depot’ and the other for pick-up by the crane, each using complementary DNA strands. To enable the crane to detach the load from the depot and lift it free, Gaub’s team made use of the fact that shear forces can easily split up DNA double helices. The pulling power of the crane acted at right angles to the helix holding the load down, peeling it apart.
On depositing the load in the target area, however, the balance of forces was decided by the different length of the two double-helical section - the double helix holding the load to the crane was designed to be shorter than the double helix attaching the load to the target, making sure that the hook of the crane could be pulled free. Using this device, the researchers wrote the letter M with 400 fluorescent molecules, achieving a target precision of around 10 nanometres.
The forces involved in DNA strand pairing can also be harnessed to create long contractile fibres much like a muscle fibre, as Daniel Lubrich’s team at the National University of Singapore have shown. Lubrich’s system is powered by ’DNA fuel’, with passive short ’staple’ sequences driving the motion of a long DNA strand. Adding the staple strand pulls closed a ’hinge’ section of the long DNA fibre, contracting the ’muscle’ - but adding the complementary strand detaches the staple, and the fibre relaxes. The waste double strand can be separated and recycled.10
Recognition
Apart from its structural and information properties, DNA can also be used as highly specific binding unit, comparable to protein antibodies. Researchers have identified many so-called DNA aptamers by selecting for binding ability from a large pool of random sequences.
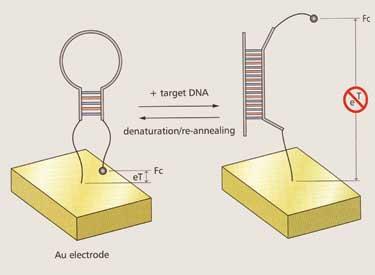
Kevin Plaxco and his group at the University of California at Santa Barbara, US, as well as a few other teams, have successfully harnessed the recognition power of DNA aptamers in highly sensitive electronic biosensors, using electrochemical signals triggered by changes in DNA conformation. Plaxco’s group has produced a gene sensor (E-DNA) as well as sensors for medically relevant proteins such as thrombin, and forensically relevant small molecules such as cocaine (see Chemistry World , July 2006, p48).
’A key advantage of these DNA-based sensors,’ Plaxco explains, ’is that they are robust, portable and so selective that they can be used directly in unmodified fluids such as blood, or environmental water samples.’ Most recently, his group has developed a sensor system that can specifically detect interactions between proteins and small molecules.11
Further forward
So with all this progress in fundamental methods, where does DNA nanotechnology stand today, and is it any nearer to real-world applications? Ned Seeman remembers that ’our laboratory was largely alone in structural DNA nanotechnology until about 2000’. Now, he estimates that there are around 40 laboratories active in the field. ’The growth of the field is still accelerating, its impact is broadening, and the excitement is extraordinary,’ he adds.
Munich, Germany-based researcher Friedrich Simmel, who has recently developed architecture-switchable materials based on DNA,12 is equally enthusiastic: ’DNA-based nanotechnology has seen an explosion of activity in the last few years with a number of impressive achievements, which make it the most advanced "technology" based on self-assembly to date apart from life,’ he told Chemistry World. ’Improvements in DNA-based biochemical circuits promise to revolutionise molecular information-processing with potential applications in life sciences and synthetic biology.’

Lee Cronin, who studies complex chemical architectures at the University of Glasgow, UK, is more cautious. ’DNA is the only polymer structure that can be configured to form reliable supramolecular assemblies, so from that point of view it is very exciting,’ he admits. ’However, I’m not sure whether any applications will come out of it beyond biology. But within the life sciences, the dynamic nature of DNA could lead to all kinds of things, even including artificial life forms.’
Perhaps the clearest indicator of the field’s potential is interest from industry. Markus Fischer from German company Entelechon, which specialises in DNA synthesis but is not involved in DNA nanotechnology so far, is optimistic about the future of this field. ’In my view, DNA is an extremely exciting material for nanotechnological applications, and I anticipate that the complexity of DNA-based nanostructures will soon exceed that of structures produced by conventional means like lithography,’ he told Chemistry World . ’Machines and dynamic structures like those already realised with DNA would certainly be much more difficult to make with other materials, especially as DNA can now be produced with high efficiency and precision, and at low cost.’
In terms of applications that could be realised in the near future, the DNA hybridisation assay on an origami nanochip looks like something that could become useful in biological research and medical diagnosis fairly soon. Even further advanced are the aptamer-based sensors. ’We are currently in negotiations with commercialisation partners,’ says Kevin Plaxco, and other researchers told Chemistry World they are already looking to patent their discoveries.
So perhaps DNA nanotechnology really is coming of age.
Michael Gross is a science writer based in Oxford, UK
References
Nature , 631 (DOI: 10.1038/350631a0) Science , 1021 (DOI: 10.1126/science.7973651) et al Nature , 423 (DOI:10.1038/nature02551) Nature , 297 (DOI: 10.1038/nature04586) et al Nature et al, Science , 180 (DOI: 10.1126/science.1150082) et al, Nature , 549 (DOI: 10.1038/nature06560) et al, Nature , 553 (DOI: 10.1038/nature06508) et al, Science , 594 (DOI: 10.1126/science.1151424) et al, Angew. Chem. Int. Ed. , 7026 (DOI: 10.1002/anie.200800476)11 K Cash et al, J. Am. Chem. Soc in press et al, Small , 1688 (DOI: 10.1002/smll.200700366)






No comments yet