Escherichia coli has been engineered to thrive on methanol not sugars. This change is the latest step toward carbon-neutral bioproduction and the synthetic strain can produce four precursor compounds used in industry.
Using E. coli to synthesise compounds such as insulin is not new and bioproduction, where the cells convert carbon into other materials, is relatively clean compared to methods using petrochemicals. However, fuelling the bacterial factories is still a problem. ‘Most of these industrial biotechnology processes are based on sugars,’ says Steffen Lindner-Mehlich, a biochemist at the Charité University Hospital in Berlin who wasn’t involved in the new research. Growing vast amounts of sugar for biorefineries is not that sustainable and diverts resources from other crops. Methanol, however, is an attractive alternative because it is sourced sustainably from agricultural waste or even carbon dioxide removed from the atmosphere. Harnessing naturally occurring methylotrophic bacteria for this purpose is tricky as these species are harder to genetically engineer compared with E. coli. E. coli is probably the most studied bacterium on the planet and is already used for bioproduction. But it can’t make use of methanol.
According to Trygve Brautaset at the Norwegian University of Science and Technology this is a surprisingly difficult problem to solve. Genome mapping shows that E. coli already possesses many of the genes and enzymes that natural methylotrophs use, but having these isn’t enough. As Lindner-Mehlich explains the two types of bacterium use the enzymes in very different ways. For molecules like sugars the pathway is linear and carbon compounds are broken up and rearranged. Methanol, with only one carbon, requires a fixation cycle. ‘It has to run three times in order to make one compound which has three carbon atoms,’ says Lindner-Mehlich. The complexity of the mutations that control the enzymes is too much for scientists to predict so they turned to evolution.
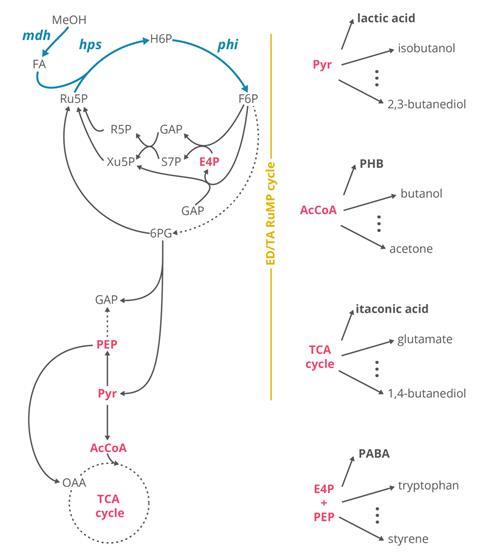
Lead author Julia Vorholt at ETH Zurich says the first step was to get E. coli ‘addicted’ to methanol. ‘If you make a mutation in a certain gene then [E. coli] needs to make a little bit of biomass for some specific compounds from methanol,’ she explains. Leaving the bacteria to grow in a bioreactor with just enough carbon to survive and an abundance of methanol favours those that can use alcohol. Natural selection takes over and bacteria which thrive using methanol outcompete the others until eventually E. coli has evolved the same fixation cycle seen in other methylotrophs.
Then the team could go back and map mutations that allowed this switch and devise ways to put it to use. ‘What we try to do is basically scan through metabolism now and look at different branch points away from metabolism towards products,’ says first author Michael Reiter. They were able to produce four products each from a different metabolic branch point and each with large markets for polymer production: lactic acid, polyhydroxybutyrate, itaconic acid and p-aminobenzoic acid. According to Reiter experiments in a commercial bioreactor showed that the synthetic E. coli was already quite capable. ‘We don’t need to do any further optimisation to just be on a level with an E. coli that utilises glucose to produce one of our products, for example, lactic acid,’ he says.
Brautaset believes that to be competitive the yields must be higher still but given the genetic tools available for bioproduction with E. coli this is a manageable problem. The doubling time of the bacteria is another area for improvement. Currently, this methylotrophic E. coli doubles every 4.3 hours at 37ºC. Ordinarily E. coli can grow almost five times as fast and so do natural methylotrophs at their preferred temperature of 50ºC. Achieving these levels at the more commercially friendly temperature of 37ºC is the next challenge, Brautaset says.
To optimise the E. coli further the Vorholt group are searching for the minimal set of mutations required for methylotrophy to make the system more robust. ‘In the future we will be able to produce chemicals hopefully with substantially reduced, maybe even zero carbon footprint,’ Reiter says.
References
MA Reiter et al, Nat. Catal., 2024, DOI: 10.1038/s41929-024-01137-0










No comments yet