An international team of researchers has used time-resolved ultrafast crystallography to follow the progress of DNA repair by a photolyase enzyme. The work is ‘the first structural characterisation of a full enzyme reaction cycle,’ says Manuel Maestre-Reyna, who led the research.
While many of the stages of this process have been studied before, the new research goes significantly further ‘by visualising the choreography of both substrate and the enzyme,’ says molecular biologist Aziz Sancar of the University of North Carolina School of Medicine, who was awarded the 2015 Nobel Prize in Chemistry for his work on mechanistic studies of DNA repair. In particular, the study has overcome the challenge of capturing events that occur at vastly different timescales to map every enzymatic step of the process. Sancar calls it ‘outstanding work, pushing the limits of time-resolved crystallography’.
‘Enzymes are slow’
Photolyases repair DNA damage caused by ultraviolet light in bacteria, fungi, plants and some animals including marsupials. Humans and other mammals don’t contain these enzymes, but we too incur light-induced damage. One common outcome is the formation of cyclobutane pyrimidine dimers (CPDs), where two adjacent pyrimidine bases (thymine or cytosine) fuse together via a four-membered cyclobutane ring. ‘CPD formation is the main cause of skin cancer, and sunburnt skin always contains CPD lesions’, says Maestre-Reyna, a biochemist at the Institute of Biological Chemistry in Taipei, Taiwan.
The enzyme repairs DNA by holding the CPD in its active site, while a coenzyme flavin adenine dinucleotide (FAD) transfers an electron to the cyclobutane ring in a process that is itself light-stimulated. This triggers a free-radical reaction that cuts the two carbon–carbon bonds holding the pyrimidines together.
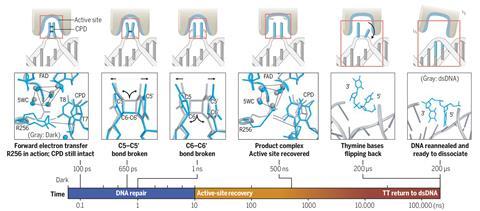
All this happens fast once FAD is activated: the initial electron transfer happens after 100ps, and the second C–C bond breaks after about 1ns. But then it takes about 500ns for the enzyme’s active site to return to its initial state, and a further 200μs for the repaired pyrimidines to flip out of the active site and the DNA to be released.
‘Enzymes are slow’, says biophysicist Marius Schmidt of the University of Wisconsin-Milwaukee. ‘Catalytic cycles complete on the millisecond timescale, yet the fundamental events such as bond formation and local relaxations are extremely fast. These two timescales are difficult to bring together.’
To follow the entire process, Maestre-Reyna and his coworkers performed ultrafast crystallography on co-crystals of a microbial photolyase and CPD-containing DNA, using two free electron laser (FEL) sources of bright x-rays. One team, working in Switzerland, collected data for the first 10 ns of the reaction, while the other team, using a FEL in Japan, studied the relaxation of the enzyme complex and release of the DNA from 10ns to 200μs.
We had to purify the protein, activate it, co-crystallise it, harvest the crystals and collect the data within 20 hours
The main challenge, says Maestre-Reyna, was that the teams had to work fast to collect the data. ‘Photolyases are only active in their fully reduced form, so all the experiments had to be performed under oxygen-free conditions,’ he says. And because the reduced form is so readily oxidised, they estimated that they had only about 20 hours before it became deactivated again. ‘We had to purify the protein, activate it, co-crystallise it, harvest the crystals and prepare them for data collection, and then actually collect the data, all on site within no more than 20 hours,’ he adds.
Maestre-Reyna stresses that the work was only possible with a large multi-disciplinary team. This included Ming-Daw Tsai at the Taiwan lab who has worked extensively on the structural basis of DNA repair, Lars-Oliver Essen of Philipps University Marburg (‘one of the most important photolyase scientists in the world’), Junpei Yamamoto of Osaka University (‘possibly the only synthetic chemist in the world capable of producing photodamaged DNA at the enormous scales required’), and Antoine Royant of the University of Grenoble Alps, an expert in performing spectroscopy in crystals.
A key, but hitherto little-understood, component of the reaction is a cluster of five water molecules within the active site. This cluster, hydrogen-bonded to a part of the protein, seems to fine-tune the active site’s affinity for CPDs and allows it to reorganise quickly when the initial electron transfer occurs – a previously speculative idea that the new work confirms. Maestre-Reyna also says that ‘while we went in armed with very good knowledge of what to expect during the first nanosecond or so, what would happen later [when the bases are released] was very much uncharted territory.’
References
M Maestre-Reyna et al, Science, 2023, 382, eadd7795 (DOI: 10.1126/science.add7795)












No comments yet