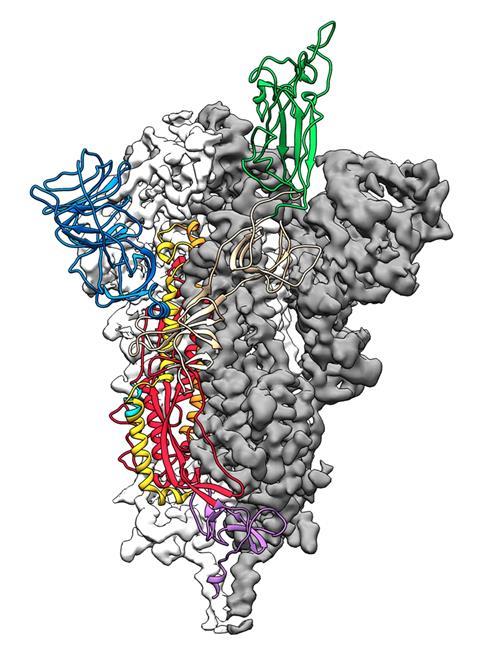
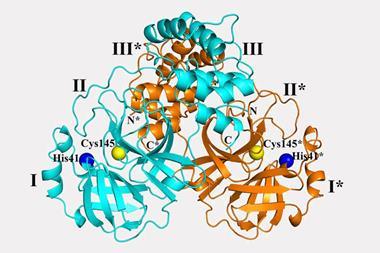
The viral spike protein on the coronavirus SARS-CoV-2 has been mapped in 3D at an atomic scale using cryo-electron microscopy. Mapping it is an essential step to developing vaccines and antiviral drugs against the virus, so the details were published at breakneck speed.
‘The spike is what we want to try and target with vaccines, with antibodies and with small molecules, so that we can prevent the virus from entering cells,’ says Jason McLellan at the University of Texas, Austin, who led the research.
The structural study has revealed that the receptor-binding domain (RBD) of SARS-CoV-2 is different to the first SARS virus – SARS-CoV-1 – from 2003–04. ‘It still retained enough [amino acid] residues so that it can bind the ACE2 receptor, but it changed other residues, maybe to evade the immune response,’ says McLellan. His team mapped out the three receptor-binding domains on each spike, which attach to ACE2 receptors. Once bound, the spike changes shape to allow the coronavirus into its target cell.
The spike differences on SARS-CoV-2 may influence how contagious it is and also affect vaccine development for the novel virus, which first emerged in Wuhan, China, at the end of 2019. There have been over 75,000 cases of the coronavirus-caused disease Covid-19 to date, and more than 2000 deaths.
McLellan’s group in recent days emailed spike protein details to about 30 different labs, as scientists race to develop a therapy or vaccine. ‘Some groups are planning drug screens, to try to identify small molecules that can bind to the spike and prevent its function,’ says McLellan. Other groups, he says, plan to use the map to develop better vaccine antigens based on the spike.
Three monoclonal antibodies that bound the first SARS virus were tested but none bound the SARS-CoV-2 virus, the study reports. McLellan says constructs from SARS-CoV-1 are likely to be ineffective for vaccine development.
A study from a Chinese group, however, published on the non-peer reviewed preprint server bioRxiv, shows a SARS-CoV-1 antibody binding potently to the spike protein of the novel coronavirus. ‘That shows cross-reactivity exists for some antibodies,’ says Peter Hotez, a microbiologist at the Baylor College of Medicine in Houston, Texas.
The SARS-CoV-2 virus binds 10 to 20 times more tightly to the ACE2 receptor than the original SARS coronavirus. ‘We don’t know the effect of that yet, but perhaps it could improve the transmissibility of the new virus,’ says McLellan. The spike protein is around 75% to 80% similar to the SARS-CoV-1 spike.
Mapping this spike structure is a critical achievement on the path to making coronavirus vaccines with greater efficacy and breadth, comments virologist Michael Farzan, at the Scripps Institute, Florida, who first reported ACE2 as the target receptor of SARS-CoV-1 in 2003. ‘The most efficient way to stop a virus is to prevent it binding to the receptor,’ he says. ‘So you are looking for antibodies that bind [the receptor-binding domain].’
‘This paper underscores the commonalities and differences between the two coronaviruses,’ says Farzan. A pan-coronavirus vaccine, to protect against all SARS-like illnesses, and to anticipate the emergence of similar viruses will eventually be needed, he adds. ‘Having two structures would help us focus on those things in common and therefore targetable.’
The first useful structure of the SARS-CoV-1 epidemic virus was completed in 2005, two years after the epidemic, and that was just the receptor-binding region. ‘The current structure was solved a month or so from the publication of the genome. This is remarkable,’ Farzan says.
References
D Wrapp et al, Science, 2020, DOI: 10.1126/science.abb2507












No comments yet