Researchers studying virus receptor-binding domains suggest virus could have passed directly from bats to humans
A structural snapshot of Sars-CoV-2 as it first touches a human cell reveals that it grips tighter than the virus behind the Sars outbreak in 2003–04.
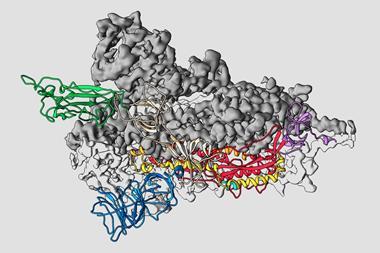
The spike proteins, which give coronaviruses a crown-like appearance on electron micrographs, first touch cells with a receptor-binding domain. In Sars-CoV-2, this docks onto angiotensin-converting enzyme 2 (ACE2) and initiates viral entry.
‘Essentially this is a picture at the molecular level of the interaction between the virus and the human cell that starts the process of invasion,’ says David Aragão, a structural biologist at Diamond Light Source, the UK’s national synchrotron facility. He uses the analogy of ‘the kiss that the virus does to seduce the human cell before using it for its own selfish purposes’.
The binding domain is a target for vaccine and antiviral drug developers. For Aragão, most noteworthy is that the results ‘show a higher affinity of the Sars-CoV-2 receptor-binding domain to human ACE2 by plasmon resonance and the structure looks to explain why it is like that.’
‘The new coronavirus has evolved new strategies to bind to its human receptor, resulting in tighter binding,’ explains Fang Li at the University of Minnesota, US, who led the research, which compared Sars-CoV-2 to Sars-CoV, the original Sars virus.1
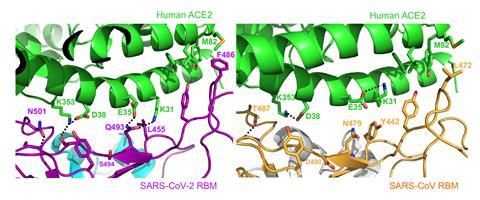
The binding domains of the two viruses form a gently concaved surface with a ridge on one side. This attaches to the outer surface of the claw-shaped ACE2 receptor in humans. Sars-CoV-2 forms a larger binding interface and touches against the receptor at more points than Sars-CoV.
Hotspots
The binding region on human ACE2 contains two positively charged and relatively large amino acids. These amino acids and their neighbours are dubbed hotspots, and must be accommodated by the virus.
‘If the hotspots are not properly accommodated, the two critical amino acids would present physical and electrostatic interferences with virus binding,’ Li says. ‘Once properly accommodated, the two hotspots contribute a significant amount of energy to virus–receptor interactions.’
‘Sars-CoV-2 accommodates the two virus-binding hotspots on human ACE2 better than Sars-CoV does,’ continues Li. ‘One region of the Sars-CoV-2 receptor-binding protein gets closer to human ACE2 and forms more extensive interactions with human ACE2.’
In a separate study, a team from Tsinghua University in China has also described the Sars-CoV-2 binding domain and ACE2 receptor.2 It suggested that similarities in the structure and sequence of the two Sars viruses were not due to them being closely related, but because of convergent evolution, with both evolving similar solutions to bind human ACE2.
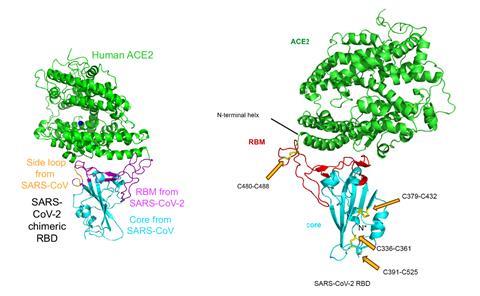
Li highlights that the Tsinghua University study agrees with the one from his team on the receptor-binding domain structure. ‘This consistency means that other researchers can confidently build on our findings when designing new drugs and vaccines to treat Covid-19.’
The Tsinghua University study ‘is useful because it shines light on the key differences between Sars-CoV and the current outbreak in the first step of infection and highjacking of human cell machinery for viral replication,’ says Aragão.
Essentially this is a picture at the molecular level of the interaction between the virus and the human cell that starts the process of invasion
‘Targeting the Sars-CoV-2 receptor-binding domain with, for example, neutralising monoclonal antibodies, could possibly stop the infection in its tracks and prevent replication,’ adds Aragão.
Li’s group plans to use the structural insights to develop antibody drugs and vaccines that specifically target how Sars-CoV-2 binds to its receptor.
Bat receptor
The most closely related virus to Sars-CoV-2 for now is a bat virus, RaTG13, recovered from a bat in China, with which it shares 96% of its genetic sequence. Structural analysis of its binding domain by Fang’s group reveals it too binds human ACE2, suggesting it can infect people. The group posit, therefore, that there does not need to be an intermediate species that passed the virus between bats and people to initiate the pandemic.
‘That RaTG13 recognises human ACE2 less well than Sars-CoV-2 does suggest that RaTG13 does not infect humans as efficiently as Sars-CoV-2,’ adds Li. For coronaviruses, receptor binding is one of the most important determinants of the viral infectivity, pathogenesis and host range.
‘RaTG13 is pretty different from Sars-CoV-2,’ says Ralph Baric, a coronavirus researcher at the University of North Carolina, Chapel Hill in the US. ‘We probably need an animal strain closer in sequence to Sars-CoV-2 to determine the mechanism of cross-species transmission.’ Structural biologists have ‘a special licence in science to make intuitive leaps that have not been proven,’ he quips. ‘[RaTG13] can bind ACE2, but that is just the first step in entry.’
References
1 J Shang et al, Nature, 2020, DOI: 10.1038/s41586-020-2179-y
2 J Lan et al, Nature, 2020, DOI: 10.1038/s41586-020-2180-5









No comments yet