Life’s molecular origins might not be preserved in the fossil record but, as Laura Howes finds out, chemists are working to fill in the gaps

Where do we come from? How did we get here? These questions have preoccupied humanity since our earliest civilisations. But answering these questions is no longer just the preserve of priests and philosophers; chemists are also addressing the problem. And in doing so they might also completely redefine the concept of life.
Back in the 1880s, Charles Darwin discussed how a ‘warm little pond, with all sorts of ammonia and phosphoric salts, light, heat, electricity, &c.’ could give birth to compounds that could evolve into life. Since then, many chemists have tried to make their own versions of that warm little pond. The most famous is the experiment carried out by Stanley Miller during his doctoral studies with Harold Urey at the University of Chicago in the US in 1953. The experiment simulated conditions believed to have predominated on the primeval Earth, including an ocean, a reducing atmosphere and a spark discharge meant to simulate lightning. The experiment created a mixture of organic molecules and amino acids, the building blocks of proteins, but Miller never succeeded in showing how that initial soup became life.
Today, labs around the world are still working on the problem of life’s origins and it will not be a short search. More than once, those who spoke to Chemistry World compared work on the problem as comparable to particle physics’ search for the Higgs particle. ‘We know more about the origin of the universe and the origin of mass than we do the origin of life,’ stresses Lee Cronin of the University of Glasgow in the UK.
Are these the polymers you’re looking for?
Imagine a graph, says John Sutherland from the University of Cambridge, UK, with chemical complexity as the x-axis and ‘aliveness’ as the y-axis. The search is to work out how you can get from the simple molecules close to where the two axes meet, to the point in the top right with the machinery of life. Do you go up in one smooth line, or are there steps along the way, and what are those steps? And what is the start point?
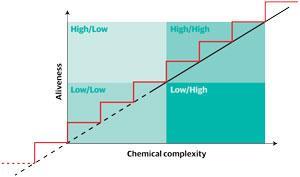
For many, the leading theory has been one referred to as the RNA world hypothesis. A phrase originally coined in 1986 by Walter Gilbert,1 who won the Nobel prize in chemistry in 1980 with Frederick Sanger and Paul Berg for work on nucleic acids. ‘One can contemplate an RNA world,’ he wrote, ‘containing only RNA molecules that serve to catalyse themselves.’
RNA is ribonucleic acid, of course: a polymer of nucleotides linked by a backbone of ribose molecules. Cells use it for many vital roles, including coding and uncoding genes and translating them into proteins. Gilbert’s optimism was born from the recent discovery of RNA’s catalytic activity. If the polymer can be used as an enzyme and information storage, people supposed, perhaps its multiple functions indicate that life began with only RNA. Proteins and DNA came later.
That idea, however logical, might be a false assumption, says Sutherland. ‘Breaking it down into “Is it an RNA world or metabolism?” or whatever was one of the most terrible things for the subject that there could have been.’
Modern biology, says Sutherland, needs compartmentalisation, metabolism, a method of storing and retrieving information, and catalysts. In the past, he suspects that people thought they were making it simpler by disconnecting these different aspects of biology, ‘but in fact I think they were making it harder’.
Sutherland’s work has been focusing on the bottom left-hand corner of his graph, at low complexity and low ‘aliveness’, trying to make the building blocks of RNA from simple molecules. As he has pieced together those first simple sugars, amino acids have also resulted from the same reactions. In fact, argues Sutherland, his work suggests that the machinery of life is predetermined by chemistry.2 The ‘inevitable reaction pathways’, resulting from the reducing chemistry possible in early world conditions is why, he says, so many biomolecules are chemically connected. ‘You can draw a proto-metabolic network which makes 12 amino acids, the nucleotides you need for RNA and some components for lipids all starting from hydrogen cyanide and a couple of other starting materials using hydrogen sulfide and light.’ Biology uses both RNA and proteins, he says, because that is what you get.
The next step, which is hardly trivial, is showing how these connect up into polymers, or at least shorter oligomers. That still will not mean molecular life, but it will be another step along the graph.
Digging down
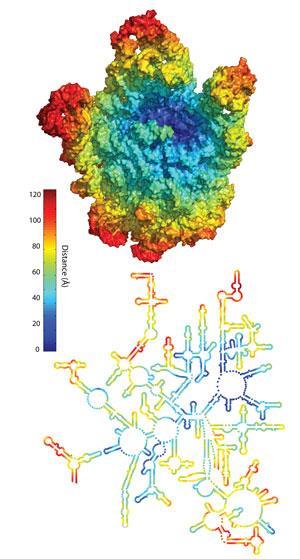
If Sutherland is working from the bottom up towards complexity, Loren Williams of Georgia Tech in the US is working down from the ‘top’. Williams’ work is mainly focused on the ribosome; made from RNA and protein chains, the large piece of chemical machinery is today responsible for protein biosynthesis. Although protein strands today help form the shape of the ribosome, none of them are directly involved in its translational work. In fact, says Williams, the ribosome is itself something of a molecular fossil.
‘The nice thing about the ribosome,’ he adds, ‘is that it’s old, older than the biology around it.’ Williams suggests that the very centre of the ribosome might be as much as 3.8?billion years old. ‘We have a model,’ Williams explains, ‘that the ribosome evolved like an onion, building layer on layer.’ As each new layer was added, he suggests, the previous layer could no longer be altered. By comparing the ribosomes of different life forms, Williams and others have shown that the central core of the ribosome is fixed, and that, he says, is the point of origin for the ribosome.
Williams does not think, however, that the ribosome was originally doing anything as complicated as creating protein. That core, he suggests, was more of a non-specific catalyst – he uses the phrase ‘molecular sausage maker’ – for different condensation reactions. Recently, Williams showed3 that in the oxygen-poor environment of early Earth, early ribosomes could have used more reactive metals, such as iron, at their centre rather than magnesium and that this increases the capability of the catalyst, so perhaps the core remained but the metal changed.
Talking about evolution
If Sutherland is trying to build up life from the very simplest of starting materials and Williams is trying to dig down to find out what the first ribosomes looked like, others are working somewhere in the middle.
Irene Chen at Harvard University in Cambridge, US, for example, is exploring what she describes as the ‘fitness landscape’ of molecules – a way of picturing the relationship between sequence and ‘fitness’, or biochemical activity. ‘Knowing the topology of the landscape is extremely important to understanding natural selection at a quantitative level, and we can actually do this for short RNA molecules,’ she explains. But her group also looks at membrane vesicles and viruses, non-living chemical systems that are ‘life-like’.
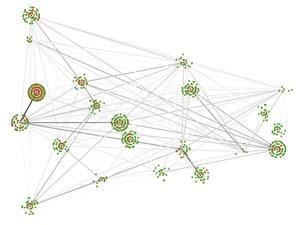
Chen’s lab group are using a combination of some modelling and a lot of wet chemistry to map these landscapes,4 and understand how small changes can affect molecular evolution. They are extending an interest in the relationship between chemical structure and biochemical activity that Chen says has fascinated her since her undergraduate days. ‘It turns out,’ she adds, ‘that this problem is very relevant to the origin of life, when there must have been a high degree of exploration in chemical structure and sequence space.’
That comment is key. When trying to understand how life on Earth emerged it is easy to think of a simple linear sequence, especially since the clues left behind are from the successes that led to life today. Just as we might say history is written by the winners, the clues left behind for chemists to follow are from the successful pathways. In between there will have been false starts, dead ends and huge amounts of molecular competition. Much of Chen’s work is in understanding and quantifying all the evolutionary possibilities, she explains, and working out the probability of different functions arising spontaneously.
However, with a question as big as the origin of life, it is unlikely that one team will find the answer on its own. ‘I think the chemistry/biology transition must be broken down into smaller chunks,’ says Chen. For her, those chunks include the appearance of templating, replication and compartmentalisation.
Luckily, there are many groups working on different puzzles along the way from Darwin’s warm little pond to life as we know it today. Both Chen and Sutherland are in part funded by the Simons Foundation collaboration on the origins of life.5 In total, 15 investigators are being funded to answer basic questions about how early prebiotic chemistry led to the earliest life, and indeed, what would that life have looked like? Others, such as Williams, get their funding from Nasa.
Ultimately, chemists can only suggest likely pathways. Without time travel, how exactly life first began on Earth will remain a mystery. But Sutherland is convinced that at some point in the future chemistry will be able to explain the basic structure and steps. ‘Finding that chemistry has been an incredibly tough task,’ he says, ‘but I do think we’ve partly found it.’
Definition’s what you need
As you look along Sutherland’s graph the question is not just how the two points are joined up, but at what point life itself can be said to exist. Today, biologists define organisms as alive if they can fulfil several criteria, including self-replication – which is why viruses are not said to be alive because they need to use another organism to replicate.
My definition of a minimal life is an autonomous chemical entity that can evolve
Lee Cronin
Systems chemistry and the study of simple self-replicating systems suggest that it is the self-replication that is key, according to Addy Pross of Ben-Gurion University of the Negev in Israel. Even before you get to what you might recognise as life, the simple molecules that led to more complicated ones would have had to replicate themselves, perhaps in cooperation with other molecules. Biology, says Pross, is just a complex kind of replicative chemistry.
Cronin defines life similarly, but with an important caveat. ‘My definition of a minimal life,’ he says, ‘is an autonomous chemical entity that can evolve.’ Evolution, he points out, requires replication but it adds another level.
Much of Cronin’s prior work has been as an inorganic chemist, but the fundamental properties of life are ones, he says, that he says he has been fascinated with for years. First, Cronin and his lab made polyoxometalate compounds self-organise and -assemble. The group extended this to begin work on the start of inorganic biology by showing how template directed synthesis can create inorganic cells by exploiting liquid–liquid interfaces.6 The next step, Cronin says, is to combine this with an understanding of systems chemistry to start designing chemical systems with the same properties that we consider unique to biological molecules. Especially that requirement for evolvability.
A a lot of his recent work on printing custom reactionware, for example, or developing a chemical reaction network, is part of what Cronin sees as a larger project to help probe how chemistry can evolve into biology. That goes beyond RNA and DNA to make a non-biological life form. ‘If I can show that evolution is the guiding principle for the establishment of a living system it will, by definition, show where life came from on planet Earth, in fact it will trivialise it in some respects.’ That is a bold claim, but certainly synthetic biology may uncover tricks that help in the search to discover how we got here, as well as potentially creating minimal lifeforms.
The chemical origin of life is a hugely important question for humanity, but not just for explaining how we got here. It will help us look for life in space, and can only lead to a better understanding of how our bridging science links physics with biology. As Sutherland says, in this respect, chemistry is the life science, and there are still plenty of secrets to uncover.
Plenty of room in the middle
Another researcher working in the ‘middle’ is Phil Holliger, who works at the UK Medical Research Council’s Laboratory of Molecular Biology in Cambridge, focusing on particular steps along Sutherland’s graph, such as making an RNAzyme that can make catalytically active RNA. Today, RNA is made by a protein-based enzyme called RNA polymerase. Before enzymes, the RNA world hypothesis suggests that RNA would have had to make itself. Holliger has been working to show that that is possible using RNA sequences that have been suggested as common ancestors to those today.
After taking an artificial RNAzyme from others’ work in the 1990s, Holliger’s group introduced evolutionary pressure to selectively improve the enzyme, and then combined the best bits of their experiments to make a new RNAzyme that replicates faster and more reliably than previous attempts. As well as making longer strands of RNA, the RNAzyme can also make enzymatically active RNA molecules, a huge step towards showing that catalytic RNA can make itself can evolve.
Holliger’s work on RNAzymes has also led him to suggest that life did not start in warm primordial pools at all. Instead, he has suggested, life on Earth had chilly beginnings. RNA, he points out, is an odd choice of molecule because it is hard to understand how it would have survived on early Earth without degrading. Holliger suggests that in cold pockets of water in ice, RNA might not have reacted quickly but would have been more stable. Using an artificial ice column, his group showed that the ice network helped concentrate the solute concentrations into small areas and that a small RNAzyme, R18, could self catalyse at low temperatures.
References
1 W Gilbert, Nature, 1986, 319, 618 (DOI: 10.1038/319618a0)
2 D J Ritson and J D Sutherland, Angew. Chem. Int. Ed., 2013, DOI: 10.1002/anie.201300321
3 C Hsiao et al, Nat. Chem., 2013, 5, 525 (DOI: 10.1038/nchem.1649)
4 J Derr et al, Nucl. Acids Res., 2012, 40, 4711 (DOI: 10.1093/nar/gks065)
6 G J T Cooper et al, Angew. Chem. Int. Ed., 2011, 50, 10373 (DOI: 10.1002/anie.201105068)











No comments yet