A strain of Escherichia coli (E. coli) with a synthetic genetic code comprising just 57 codons, rather than the standard 64, is the most significantly recoded organism to date. The new strain, named Syn57, demonstrates that life can function with a significantly compressed genetic code.
The researchers who carried out the work are based at the Medical Research Council’s Laboratory of Molecular Biology (LMB) in Cambridge, UK. They say that by freeing up codons – sequences of three nucleotide bases that correspond with specific amino acids – in the E. coli genome, Syn57 has more space to introduce unnatural amino acids. This could open up new applications, such as generating organisms that are resistant to viruses or produce new enzymes.
The same team previously made Syn61 – a strain of E.coli with 61 codons – in 2019. But the researchers were keen to find out if living organisms could tolerate additional codon compression, taking them further away from their natural genetic sequence.
‘We wanted to know how deeply you could compress the genetic code, because if you can compress it, you can then free up some of the previously redundant codons to repurpose them for a new application,’ explains Wes Robertson, a synthetic biologist at the LMB and co-leader of the project. ‘The idea is we can then use cells to make things that chemists used to make in a flask, but now we can do it in a more programmable [and] bio-sustainable way.’
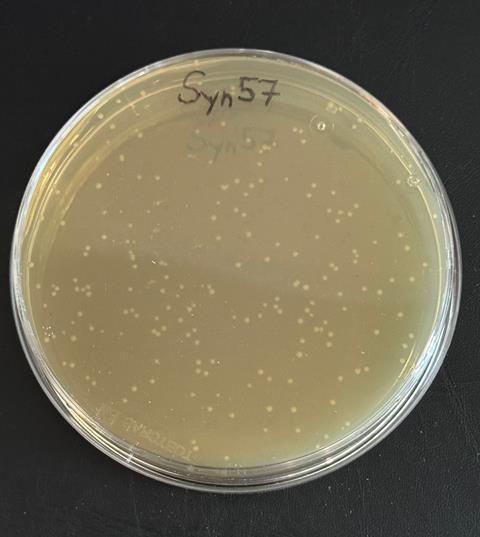
To do this, the team started by developing a recoding scheme that would free up seven codons in the E. coli genome; four of the six codons which encode the amino acid serine, two of the four codons for the amino acid alanine and one stop codon. In total, this meant making more than 100,000 codon changes across the 4 million base pair genome of E. coli.
To make the task more manageable they split the genome up into 38 fragments of around 100,000 base pairs each and synthesised them individually using homologous recombination in yeast to ensure that the recoding scheme would work.
‘It worked for about 75% of them. For the 25% where it didn’t work, we then went in and mapped, via a variety of new linkage mapping techniques that we developed,’ says Robertson. ‘Once we could pinpoint the problems, we added different synthetic DNA designs, which maintained compression, but were slightly different to our original design. ’
They then stitched the fragments together, fixing potential problems as they went to enable the next step of the synthesis.
While Syn57 didn’t grow as well as the original strain, Robertson notes that it ‘grew well enough for us to characterise it in the lab’. He adds that further modifications could provide Syn57 with a ‘genetic firewall’ that would prevent it interacting with genetic material from the wild-type E. coli. ‘So this will yield a virus-resistant strain which could be quite useful in industrial purposes,’ he says.
Encoding new chemistry
Martin Spinck, who also worked on the project, says the reassignment of the codons is limited only by their creativity. ‘All of these seven codons can be reassigned to any combination of seven or a subset of seven unnatural amino acids … and these can introduce quite a lot of new motives into biology that naturally would never exist and would never occur.’
Farren Isaacs, an expert in molecular, cellular and developmental biology at Yale University in the US, describes the construction of a genome with 57 codons as a ‘significant’ accomplishment, although, he adds that the new functions that emerge will ultimately determine the impact of the work.
‘The key aspect of the design of this genome is to open up coding channels,’ he says. ‘There’s a number of potentially very useful properties that can emerge from organisms with a new genetic code: you can repurpose those codons to encode new chemistry, to create new kinds of synthetic proteins and polymers.’
‘They can confer resistance to viruses and other forms of horizontal gene transfer,’ he adds. ‘And you can also use them to engineer novel biocontainment solutions, where you can actually engineer these organisms to be dependent on synthetic amino acids preventing growth or escape in the wild.’
‘It’s the function that emerges that I think is most compelling for synthetic genomes with alternative codes.’
However, Isaacs says there are still several questions yet to be answered. ‘What they haven’t done yet is knocked out tRNAs or release factors that decode those codons they have eliminated and see how the cell responds – does it completely eliminate that function from the cell or are there other translation factors that overlap and respond? … How might it impact growth and viability?’
‘That is going to be essential in actually realising the function of these organisms.’
References
W E Robertson et al, Science, 2025, DOI: 10.1126/science.ady4368













No comments yet