Whole blood method for diagnosing bacterial infections has unprecedented speed and sensitivity
A new test can identify bacteria and whether they bear a variety of antibiotic resistance genes within one hour. The test, which analyses whole blood and uses the polymerase chain reaction, could lower the death toll of sepsis by helping to identify the best antibiotic to administer to patients with a bloodstream infection.
The World Health Organization estimates that sepsis causes six million deaths a year. ‘Studies have shown that every hour of delay leads to increased probability of death and disability in patients,’ says Dino Di Carlo, an expert in bioengineering at the University of California, Los Angeles in the US, who was not involved in developing the new test. It is therefore critical that patients receive appropriate antibiotics as quickly as possible. Unfortunately, identifying the most appropriate antibiotic requires taking a blood sample, extracting the DNA, then culturing and analysing this sample – a process that can take several days. This is why initial treatment is usually a very broad, but potentially ineffective, antibiotic.
Now researchers at the University of California, Irvine led by Weian Zhao, have designed a PCR system that can use whole blood, meaning they do not need to culture or process the sample, something he says was impossible before. ‘It cuts out a bottleneck to get to an initial result on the presence of species-specific and susceptibility genes,’ comments Di Carlo. In addition to the massive time saving this affords, the sensitivity of this method is 10–100 times better than existing methods, credited to the three dimensional particle counting method that Zhao’s team has developed.
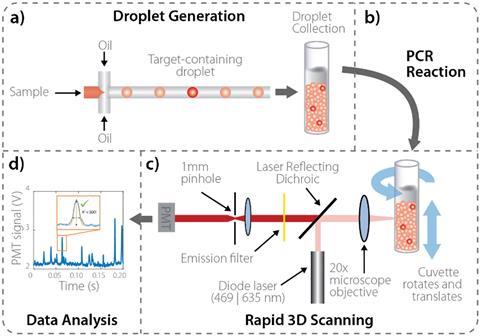
The test uses a microfluidic device that mixes the whole blood sample with PCR reagents, which work with whole blood, and then turns this mixture into millions of individual picolitre-sized droplets. Following the PCR reaction, droplets containing particular genetic targets become fluorescent and are detected and quantified by a high throughput 3D particle counter. The counter works by moving the sample up and down, and rotating it whilst it is analysed. Compared to existing methods, analysing a millilitre of liquid takes minutes rather than hours, and by analysing a higher volume of liquid it increases the chance of discovering pathogenic bacteria.
Zhao says the team’s ongoing goal is to ‘implement this technology in a clinical setting’ and says they are working on completely automating the process. Ultimately, the team is hopeful that this research will improve antibiotic stewardship and early, appropriate treatment of bloodstream infections.
References
This article is free to access until 17 March 2020
T J Abram et al, Lab Chip, 2020, DOI: 10.1039/c9lc01212e










No comments yet