Doctors and pharmaceutical companies are beginning to open their eyes to the power of mass spectrometry imaging, finds Nina Notman
Imagine what drug discovery would be like if you could look directly into a cell and see where your drug ends up. This could go some way to reducing the staggering cost of developing new drugs, by weeding out ineffective candidates before carrying out expensive trials. ‘There’s a huge push in the pharmaceutical industry to reduce attrition in drugs,’ explains Ian Gilmore from the UK’s National Centre of Excellence in Mass Spectrometry Imaging in Teddington.
The number of new drugs approved per billion dollars spent on R&D has halved roughly every nine years since 1950 (adjusting for inflation). In the 1950s, more than 50 new drugs were produced per billion dollars of investment, whereas today not even one drug can be brought to market for that amount of money. ‘If you can spot a drug is going to fail much earlier in the pipeline then you don’t waste a billion dollars trying to get it to market,’ Gilmore says.
Peering inside a cell
Many drug targets are inside cells and Gilmore’s team – in collaboration with the instrument manufacturers Ion-TOF and Thermo Fisher Scientific and scientists at the Stevenage, UK, site of pharmaceutical giant GlaxoSmithKline (GSK) – are aiming to develop a very high resolution 3D mass spectrometry imaging technique that can see where a potential drug molecule goes once it gets into a cell. It is hoped that this will allow potential problems to be identified at a much earlier stage than they can at present.
We want to be able to see if the compound gets to the target area
The team can currently tell whether a molecule has entered a single cell in sufficiently high concentrations to have the desired therapeutic effect or whether it is just coated on the outside of the cell. ‘You can also see if the drug has engaged with its target or whether a drug metabolite is present as well,’ says Gilmore. The next generation instrument – currently in development – is aiming to enhance the sensitivity by two orders of magnitude and allow researchers to see where the drug goes at the organelle level. ‘Ultimately, we want to be able to see if the compound gets to the target area within the cell,’ explains GSK’s biological mass spectrometry manager Andy West.
This resolution will also allow researchers to check that potential drug molecules are not lodging within the wrong cellular components, potentially causing toxicity. This is a fairly common problem. ‘Well-studied drugs such as [antimalarial] chloroquine build up a relatively high concentration, specifically in the sub-cellular lysosomes, and can cause toxicity,’ says West.
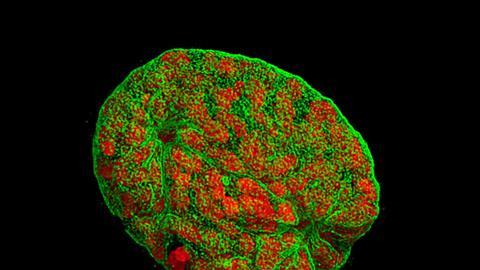
The imaging technique they are using for this is 3D secondary ion mass spectrometry, or Sims. In this technique, ions (known as primary or projectile ions) are fired at a surface in a vacuum. The bombardment causes molecules and their fragments to be released from the surface, some of which are ionised. These secondary ions are then analysed in a mass spectrometer. To obtain spatial maps in 2D, the primary ion beam is scanned across the surface and a spectrum is collected at each pixel. To get a 3D image, a second ion beam is used to shave off the surface, layer by layer. A 2D image is taken of each layer and these are then stacked on top of each other to give the 3D image.
To prepare the samples, the cells are first incubated with the molecule of interest. They are then frozen and dehydrated before being put in the mass spectrometer. ‘This is not ideal because the cell collapses,’ explains West. ‘This is something we are working on improving.’
The instrument is already being embedded into GSK’s drug discovery process. ‘We are trying it on several different things and it’s giving us some very interesting data,’ says West. Ultimately, the aim is to look at all potential drug molecules with intracellular targets using Sims during lead generation. This stage, where molecules that have given a ‘hit’ during a high throughput screen undergo preliminary optimisation, occurs near the beginning of the drug discovery process. There is no comparable analytical technique able to provide the same information at such an early stage, West says.
Moving forward
The ultra-high resolution offered by Sims is also used for other practical applications, including in the design of efficient polymeric drug delivery systems, and for fundamental biological research such as identifying new biological pathways and observing the highly-curved lipid membranes that form when protozoa mate. But it isn’t as widely used as some of the other mass spectrometry imaging techniques, mostly due to the cost of the machines. ‘They are commercially available, but they are £1 million plus,’ says Nick Winograd, an analytical chemist at Pennsylvania State University in the US. ‘Only 30 or so are sold a year.’
One of the focuses of Winograd’s group is developing new types of primary ion beams for the ionisation process. Firing atomic ions at a surface can cause the molecules to fragment so much that it is difficult to figure out what the original molecule was from the resulting complicated spectra. Cluster ions are commonly used today to overcome this, with Winograd’s favourite projectile being C60. Large argon clusters are also popular. ‘These have improved the quality of our spectra quite a bit,’ he says. He does, however, raise concerns that commercial instruments are not yet optimised for these new cluster ion beams: ‘I think we are losing resolution because of the way they are set up.’
Another issue holding back further resolution improvements, according to Winograd, is the tiny number of secondary ions that are released from the surface when it is bombarded. ‘The ionisation is really inefficient. For organic molecules, proteins and so on, it’s probably one in a million molecules that are coming off as molecular ions. If we could increase the ionisation efficiency, I think that would also be an extraordinarily happy outcome.’ Ionisation efficiency is a problem for all the mass spectrometry imaging techniques that rely on desorption, he adds. ‘We are trying a bunch of things to try and increase ionisation efficiency.’ One of the problems, explains Gilmore, is that some of the natural materials in cells, such as salts, suppress the ion signals.
The other big challenge is with the amount of data produced, according to Gilmore. ‘In a day’s analysis, you could be generating 100 gigabytes of data.’ A unique spectrum is collected for every pixel of these 3D images: ‘They are mind-boggling complicated data sets.’ Mining this volume of data for the desired information is problematic. ‘We need to use mathematical techniques like multivariate analysis to find regions of the images that are similar, and this involves some fairly heavy computational ability.’ Even with the computers we have today it’s still too slow, and better ways to analyse and search the data are needed, Gilmore adds.
Brain surgery
But not all mass spectrometry experts are hankering after better resolution and large data sets. Graham Cooks, from Purdue University, US, is instead focused on speed in his quest to get mass spectrometry imaging into the operating theatre. ‘If you’re doing diagnostics, in real-time on-site during surgery, then detailed spatial cellular level information is really not required,’ he explains. ‘Surgeons’ decisions are limited to the scale on which they work, which is on the order of millimetres and the mass spec experiment easily goes submillimetres.’
For this, Cooks is using desorption electrospray ionisation (Desi), a mass spectrometry imaging technique that was developed in his lab about a decade ago and has been used for a host of applications ranging from detecting explosives to answering fundamental biological questions. In Desi, a stream of charged aqueous spray droplets desorb and then ionise molecules from a surface. This is a far gentler form of ionisation than Sims, therefore much less fragmentation occurs and the spectra are easier to interpret. Desi is carried out under ambient conditions (unlike Sims, which requires a vacuum), but cannot produce 3D images without slicing up a sample and analysing each layer separately.
Trials are currently underway to test Desi’s viability in supporting surgeons during the removal of cancerous tumours. Questions can arise during these operations concerning the type or grade of tumour and whether all the diseased tissue has been removed, and at present the only way to get answers is to send a tissue sample to the hospital’s pathology labs. ‘I watched [an operation to remove a brain tumour] a couple of months ago and there was only one chemical analysis done. One tiny section was sent to pathology during the surgery and, 20 minutes later, the answer was telephoned back,’ explains Cooks. The pathologists look at the samples under a microscope.
For the trials, when the surgeons have a question, they do two smears between glass slides rather than one. The first goes to pathology as usual, while a second is held in front of a Desi instrument in the operating theatre. The instrument then rapidly runs the spectrum of the lipids in the tissue sample against a library of spectra that have previously been collected for that disease. From that, a ‘disease’ or ‘non-disease’ status is assigned to the tiny piece of tissue that is being analysed, explains Cooks. If the tissue is diseased, the type of tumour, grade of cancer and tumour cell concentration can all be determined by Desi based on work by Cooks and Nathalie Agar at Brigham and Women’s Hospital, Boston.1 ‘All of that information comes in the blink of an eye,’ says Cooks. This is being trialled for both brain and breast cancer at present. ‘The brain is 50% lipids, and so it makes enormous sense to do this experiment on brain tissue.’
iKnife
A past postdoc of Cooks, Zoltan Takats – now at Imperial College London, UK – is developing a similar device for use in cancer surgery based on the rapid evaporative ionisation mass spectrometry technique, known as Reims.
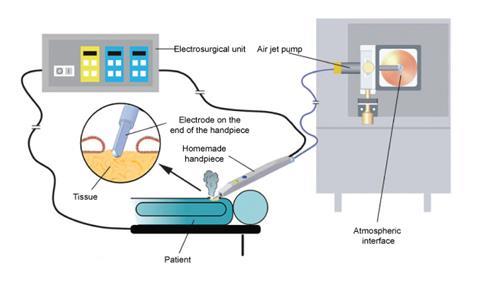
The iKnife analyses the ions in the smoke emitted as standard electrosurgical instruments cut through tissue. These knives are already fitted with a tube to take the smoke away from the operating table, and the mass spectrometer is simply fitted onto the smoke collection system. As the surgeon cuts, the spectra of the hundreds of different lipid ions in the smoke are continually collected and compared against a library. The machine then gives a continuous, real-time red or green feedback on a screen to the surgeon. ‘If it’s red, then it is the tissue which one shouldn’t cut into. The green tells the surgeon everything is all right,’ explains Takats. ‘It also gives histological information explaining exactly why it turned red.’
In 2013, the iKnife was trialled in 81 cancer tumour removal operations and the results compared to traditional microscope techniques.2 The iKnife correctly identified the tissue 100% of the time. No more trials have been carried out since then, as Takats is currently waiting for a clinical grade version of his device to be designed. A medical device has to be designed from scratch even if it is something that already exists, he explains. ‘Hopefully next year this device will be ready [for clinical trials].’
Desi at the doc’s
Cooks is hoping that small and simple Desi instruments will also find a home in doctors’ surgeries to enable the rapid diagnosis of bacterial infections such as strep throat. For most of us, strep throat – caused by Streptococcus pyogenes– is harmless and fixes itself within a couple of days, but in the elderly and very young it can lead to serious complications such as rheumatic fever. This infection can be easily treated with antibiotics, but the results from the rapid immunity diagnostic test that is currently used are not considered to be very reliable. ‘It turns out that it’s relatively easy to distinguish the Streptococci species that’s involved in strep throat from other species of microorganisms [using our instrument],’ says Cooks. And the results 3 take just 10 seconds.
Trials using this instrument are just starting in Indianapolis. A dual swab is taken from the mouths of patients suspected of having strep throat, and one half is sent for standard analysis whilst the second is analysed using the mass spectrometer. The results are then compared. ‘If the results turn out to be good, it would mean that we can have a very small, simple mass spectrometer that could detect strep throat better than is being done at the moment,’ says Cooks.
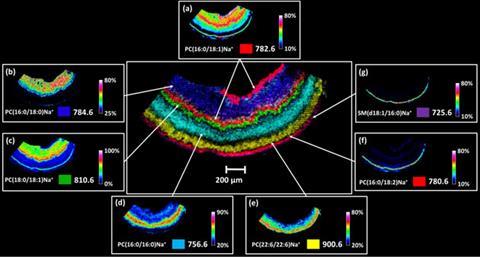
Meanwhile, chemist Richard Caprioli at Vanderbilt University in the US is championing the use of another mass spectrometry imaging technique – Maldi (matrix-assisted laser desorption/ionisation) – in the clinical arena. He has already carried out preliminary tests to demonstrate its viability in detecting fatty liver disease in human liver samples and a juvenile onset form of macular degeneration in retinal tissue, with encouraging results.4 Again, lipids are under the mass spec microscope.
For Maldi analysis, the samples are coated in a matrix before being placed in the mass spectrometer. ‘We do this robotically and therefore it is pretty straightforward and easy to do,’ says Caprioli. The molecules are then desorbed and ionised by sweeping a gentle laser beam across a sample’s surface under a vacuum. Due to the gentle ionisation method, the spectra are relatively easy to interpret, like those from Desi. Caprioli’s team then runs these against a library of spectra for the disease it is trying to diagnose. ‘We are working on protocols to make it really no more difficult than looking at a slide under a microscope,’ he says.
A combined future
Cooks, Caprioli and Takats all agree that it is just a matter of time before mass spectrometry imaging becomes an integral part of our healthcare systems. The remaining challenges are regulatory not scientific, says Cooks. ‘There’s a little doubt in my mind that, after appropriate testing and validation, this will become very important in anatomic pathology,’ says Caprioli. ‘In the years to come, pathologists will be using mass spectrometry-based images for diagnosing and prognosing disease, and less of the antibody-based techniques.’
But they are not setting out to completely replace the microscope. ‘Any imaging modality has very strong points and weak points. For example, normal microscopy can get very high spatial resolution images but there’s not a lot of molecular information in that image. Mass spectrometry is just the opposite. It doesn’t have the high resolution like microscopy, but it has exquisite mass molecular resolution,’ says Caprioli. ‘So, what we’ve done is to mathematically combine these two images into a brand new image.’
And the pricing may be attractive too. Cooks’ Desi machines are being purposely designed to be as cheap as possible. For Maldi, ‘Once the instrument has been purchased, the cost per analysis is very low,’ says Caprioli. Meanwhile, the analytical manufacturer Waters, which is currently designing the clinical grade iKnife, has an alternative approach to enticing hospitals to use the instrument. ‘Their business plan is to give the instrument to hospitals for free and then charge per intervention,’ says Takats. ‘I believe that it would be a few hundred pounds per intervention, which is negligible to the overall cost of cancer patients undergoing surgery.’
‘This really is a very exciting time for mass spectrometry imaging,’ concludes Gilmore. ‘The three principal areas – Sims, Maldi and the ambient techniques – are all developing at a very fast pace at the moment.’
Nina Notman is a science writer based in Salisbury, UK
References
1 S Santagata et al, Proc. Natl. Acad. Sci. USA, 2014, 111, 11121 (DOI: 10.1073/pnas.1404724111)
2 J Balog et al, Sci. Transl. Med., 2013, 5, 194ra93 (DOI: 10.1126/scitranslmed.3005623)
3 A Jarmusch et al, Analyst, 2014, 139, 4785 (DOI: 10.1039/c4an00959b)
4 D M Anderson et al, J. Am. Soc. Mass Spectrom., 2014, 25, 1394 (DOI: 10.1007/s13361-014-0883-2)











No comments yet