
A new antibiotic that targets only a specific strain of bacteria, while sparing friendlier bacteria, might one day bring some respite to patients with inflammatory bowel diseases (IBD). The development of this antibiotic is part of the first wave of drugs whose development has been smoothed using AI.
Most of the antibiotics in use are broad spectrum so kill both harmful and friendly bacteria, causing imbalances in gut flora. This can become a serious problem for patients with IBD, such as Crohn’s disease. Patients with these conditions can find their intestines colonised by high levels of Enterobacteriaceae, especially an invasive strain of Escherichia coli. This invasive E. coli strain replicates within intestinal epithelial cells and macrophages and is resistant, on average, to four or more antibiotics. ‘Millions of people around the world suffer from Crohn’s, roughly 30% have [invasive E. coli] infections, and these patients don’t have ideal treatment options,’ says Jonathan Stokes, a computational chemist at McMaster University in Canada.
Stokes and his team screened a diverse collection of over 10,000 molecules and identified those that halted the growth of invasive E. coli. ‘Filtering’ these molecules through a series of computational and experimental ‘sieves’ yielded a molecule that they named enterololin that does not structurally resemble any known antibiotic. They also found that using enterololin with SPR741, which disrupts the outer membrane of Gram-negative bacteria, enhanced its lethality.
Experiments with different Gram-negative and -positive bacteria showed that only enterololin targets members of the Enterobacteriaceae family, while sparing other bacteria. When combined with SPR741, its minimum inhibitory concentration (MIC) was 30ng/ml, which is substantially lower than 1µg/ml, the MIC of many antibiotics. When mice infected with this invasive E. coli were treated with enterololin and SPR741, it eradicated all these bacteria from their intestines with negligible damage to friendly microbiota. Even at higher doses the mice didn’t show any adverse effects.
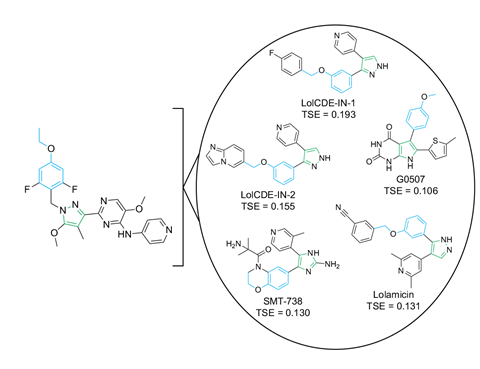
Next, the researchers employed a machine learning (ML) algorithm, a diffusion model that predicts binding poses for small molecules, to predict which protein complex enterololin targets. They found that it targets a complex that helps to make these bacteria resistant to large, hydrophobic antibiotics. Experimental work proved this prediction correct and enterololin was found to inhibit just this protein complex of the Enterobacteriaceae family, making it highly selective. The researchers estimate that this AI-driven approach saved them approximately two years of research and a million dollars. ‘Guided by ML, we [deciphered enterololin’s mechanism of action (MOA)] in six months for $60,000 (£46,000),’ says Stokes. ‘So far as I know, it’s the first time a diffusion model has been prospectively applied to help guide MOA elucidation, which is really exciting!’
Since the AI-aided discovery of halicin in 2020, there has been a flurry of novel antimicrobials that were designed or discovered by AI. What was earlier dependent on years of trial and error is being replaced by machine-learning algorithms producing novel antibiotic designs, tailor-made to target certain bacteria. Whether it’s sifting through hundreds of thousands of molecules to find a potent antimalarial, designing new antibiotics from scratch that can kill multidrug resistant bacteria or discovering an entirely new class of antibiotics, AI has helped to galvanise antimicrobial development. But so far, none of the AI-driven antimicrobials have made it through human trials and there’s still some scepticism about how useful AI will really be in drug discovery.
Olivier Espéli, a bacteriologist at Collège de France in Paris who wasn’t involved with the study, calls this work ‘particularly interesting and exciting’ and ‘encouraging’. ‘Being able to eliminate E. coli without altering what remains of the anti-inflammatory microbiota would be excellent news,’ he adds. He explains that allowing anti-inflammatory bacterium to multiply may ‘block the vicious cycle at play in Crohn’s disease’.
Maria Teresa Abreu, a gastroenterologist at Cedars–Sinai hospital in the US, who also wasn’t involved with the study, says the combination works but tests in human subjects are needed to assess its clinical potential. However, she has concerns that ‘the proof that it works in vivo is by [force feeding] mice with [invasive E. coli] and then demonstrating eradication in luminal contents of the cecum and colon’. This doesn’t address if the mice developed intracellular infection and if this combo can eradicate invasive E. coli hiding within macrophages.
References
D B Catacutan et al, Nat. Microbiol., 2025, 10, 2808 (DOI: 10.1038/s41564-025-02142-0)








No comments yet