A chemical nose developed by researchers in Japan can identify certain health conditions by analysing the microbiome of stool samples.
The gut microbiome is known to reflect several health conditions and general wellbeing, so tools to monitor its composition present an opportunity for routine observation and rapid diagnosis. However, microbiomes are complex systems, which overwhelm many existing diagnostic techniques due to the high number of biomarkers present.
Tools known as chemical noses, however, looks for patterns of biomarkers. ‘When we drink something, we don’t identify each molecule contained in the drink. Instead, we understand the pattern of the responses of several kinds of cells,’ explains Shunsuke Tomita, from the National Institute of Advanced Industrial Science and Technology in Ibaraki. A chemical nose works in the same way: ‘We understand the patterns of the responses [from the sensor], so we can detect many kinds of analytes … and understand complex systems.’
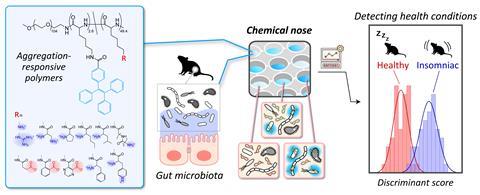
The chemical nose developed by Tomita and his colleagues uses a set of different polymers with two important characteristics: charged groups that interact with the charged membrane on the surface of bacteria, and a tetraphenylethene group that produces a fluorescent signal, which is altered by binding. Stool samples are incubated in a 96-well plate at pH5 and pH7 along with one of the polymers, and a plate reader measures the fluorescence signals in each well. Data from this array creates a pattern for each sample and by comparing those patterns to patterns of known bacterial strains, a pattern recognition algorithm can identify bacterial combinations in the sample. Importantly, very little human input is needed to generate and analyse the large quantity of data from each sample. ‘[It] can be easily automated. There are no complicated processes, so we can easily use robotic technologies,’ says Tomita.
To test their chemical nose, the researchers applied it to samples simulating key differences in gut microbiomes associated with obesity in humans. They found that the patterns between these samples were sufficiently distinct to correlate samples with varying degrees of obesity. In a further test, the researchers used real stool samples from mice to establish patterns that were associated with sleep-deprived subjects versus well-rested subjects.
‘Being able to look at gut health and gut bacteria is becoming so important as people recognise the impact that has. It’s cool that they went after [gut microbiota] – that’s a big challenge,’ comments William Peveler, who leads a group developing biomedical sensing devices at the University of Glasgow, UK. ‘They could pick out individual microbes and even mixtures in buffer with this multi-part array. Although I think the portion of the study looking at mouse health via their droppings will need more validation to show that it’s really the microbiota being measured.’ Tomita acknowledges this next step: ‘The classification accuracy is not 100% using this polymer-based approach, so we need to improve it to get more precise information from the microbiome.’
Tomita adds that healthcare is not the only application of microbiome monitoring: ‘There are many kinds of microbiome in the environment, for example soil and plants, and these microbiomes are important for improving crops or monitoring the environment. We believe this system could be used for such environmental microbiomes.’
References
This article is open access
S Tomita et al, Chem. Sci., 2022, 13, 5830 (DOI: 10.1039/d2sc00510g)









No comments yet