Researchers in the UK have identified a unique reaction pathway between two natural products made by the Streptomyces formicae KY5 bacteria that live on ants in Kenya. These compounds, a fasamycin and a formicamycin, have previously been shown to possess potent antibacterial activity against methicillin-resistant Staphylococcus aureus (MRSA). Understanding how the bacteria turn the flat fasamycins into three-dimensional formicamycins could ultimately help chemists develop new drug leads.
Formicamycins are a family of antibacterial aromatic polyketides, discovered in 2017 by Barrie Wilkinson, from the John Innes Centre in the UK, and his colleagues. ‘We isolated them from a new microbe living in fungus-farming ant systems – these are environments where microbes have evolved in competition with one another. And we have a hypothesis that it’s a good place to look for microbes that might make biologically active molecules, in particular antibiotics.’
The same microbe also makes related polyketides called fasamycins. The fasamycins are quite flat but the microbes transform them into formicamycins, which have a twisted chair-like structure. Wilkinson’s team wanted to know how one family converts into the other.
First they performed bioinformatics analysis to identify the gene cluster involved and to identify the specific gene products required to convert fasamycins to formicamycins. The team used Cas9-mediated gene editing to make deletions in forX and forY, the genes encoding the enzymes involved in the transformation. This stopped formicamycin production and chemical analysis then identified intermediate compounds in the pathway. This revealed that the reaction proceeds by a two-step ring expansion–ring contraction process.
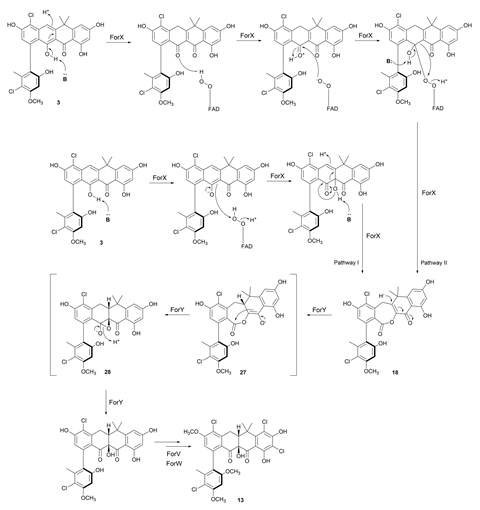
The enzyme, ForX, first catalyses a Baeyer–Villiger reaction on the fasamycin compound, which inserts an oxygen next to a carbonyl group to give a ring expanded lactone. The enzyme, ForY, then catalyses a two-electron reduction of the lactone by adding a hydride which likely proceeds via an epoxide intermediate and leading to a ring contraction. ‘The first, Baeyer–Villiger-like step is a relatively common observation in biosynthesis,’ explains Wilkinson. ‘What was really interesting was the second transformation. Although ring contractions are not that uncommon in biosynthesis, this kind of reductive ring contraction is quite unusual.’
‘This work reminds us that nature never fails to surprise us with what they can do to a chemical,’ says Thu-Thuy Dang, a biosynthesis and natural products expert from the University of British Columbia in Canada. ‘It’s very interesting to see how the enzymes are working together to orchestrate the transformation.’ Commenting on the Baeyer–Villiger reaction, Dang says ‘this type of reaction in a chemical laboratory would require strong and hazardous catalysts such as a peroxyacid, which produces waste that needs to be dealt with properly.’
‘In our quest for new antibiotics, understanding the formicamycin pathway gives us hope on the power of harnessing these pathways to combat the increasing number of new diseases that we have to deal with,’ adds Dang.
Wilkinson’s team identified two potential pathways for the initial reaction involving ForX and say that further work will investigate this and the unique reductive ring contraction in more detail.
References
These articles are open access
1 Z Qin et al, Chem. Sci., 2020, DOI: 10.1039/d0sc01712d
2 Z Qin et al, Chem. Sci., 2017, 8, 3218 (DOI: 10.1039/c6sc04265a)











No comments yet