Citizen scientists use a computer game to design brand new proteins
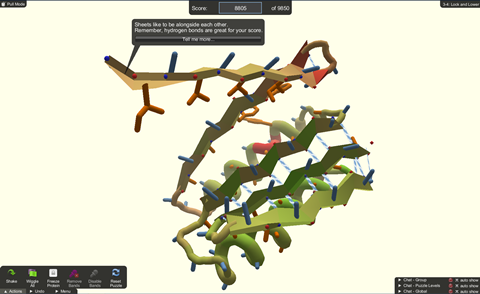
For over 10 years, members of the public have been playing a video game that challenges them to fold proteins to form the most stable structures. Developed by researchers in David Baker’s lab at the University of Washington, Foldit now has 735,000 registered users.
Early versions of the project were inspired by the Rosetta@home distributed computing project, in which people can volunteer their computers to run protein folding calculations. When the computer goes to sleep the project starts running biochemistry calculations. As it does so a screensaver pops up, showing the resulting changes in protein structure or a long polymer being folded into a three-dimensional shape. But participants wanted to be able to interact with the screensaver.
‘They were watching the computer try these different protein folds and could tell that some of them were not viable,’ says Brian Koepnick, a research scientist at the Institute for Protein Design at the University of Washington. ‘They wanted to be able to intervene and tell the computer when it was doing something good and when it was doing something bad.’
And so the idea for Foldit was born; a video game that allowed non-experts to take full control of protein folding simulations.
More than a screensaver

Through Foldit, citizen scientists have contributed to several important discoveries. In 2011, for example, players were able to correctly predict the structure of a component of the Mason-Pfizer monkey virus, which shares similarities with HIV. ‘In biology, a lot of times, if we want to understand something, we also study its evolutionary neighbours,’ explains Koepnick. ‘So, studying this related virus is very important for HIV.’
At the time researchers had measured diffraction data from a crystal of the protein but they weren’t able to ’bring it all the way home’ and solve the crystal structure. Foldit players came up with a model within two weeks of being provided with the dataset. ‘That was really amazing for the players; they did something that expert scientists had not been able to crack in 10 years,’ says Koepnick.
Unnatural designs
In recent years the focus of the Baker lab has shifted, from predicting existing protein structures to designing new proteins. ‘We’re not satisfied with the proteins that nature has anymore,’ says Koepnick. ‘Nature has some very elegant proteins that do some elegant stuff, but we want to do other stuff that native proteins aren’t built for.’ Some of this ‘other stuff’ includes designing proteins that can break down plastics, generate hydrogen sustainably, or even act as immunoproteins for newly evolved pathogens.
The research team were keen to integrate protein design into Foldit, but it was tricker than they expected. It took them about seven years to optimise their protein design software so that players could use it. The results were published in Nature last month.
The research team were keen to integrate protein design into Foldit, but it was tricker than they expected. It took them about seven years to optimise their protein design software so that players could use it. The results were published in Nature last month.1
Players unlocked their creativity by building brand new proteins. Starting with a long peptide chain, they were challenged to find an amino acid sequence that could fold into a well-structured protein, with no restrictions placed on the types of folding they could choose to use.
Researchers then created synthetic genes that encoded 146 Foldit player designs. Over a third of them were successfully expressed in E. coli and found to ‘adopt stable monomeric folded structures in solution.’ The researchers were also impressed with the diversity of the designs, with one including a brand-new type of fold that hasn’t been seen in naturally occurring proteins.
Koepnick thinks that the lack of formal scientific training amongst gamers is the reason for the variety in the structures that they propose. ‘When practising scientists design proteins, they tend to apply a lot of heuristics, simplifications, or even biases that they’ve picked up … they have very well-defined ideas about what a protein looks like.’
The research team determined high resolution structures for four of the designs and the Foldit players who designed these structures were credited as authors on the Nature paper. ‘We wanted to incorporate all of the players into the paper writing process, but in the end, we couldn’t do that for all 300 people in the consortium,’ says Koepnick. ‘At least we can do it for these four or five people who were a little more involved in the figures and the revisions.’
The Foldit project team are starting to look at other problems the gamers could help with, such as designing therapies for cancer or other future diseases, and are keen to recruit even more players to help. ‘The learning curve for Foldit is very, very steep, but Foldit players … are very excited to share what they know and get new people involved,’ explains Koepnick. ‘So new members tend to feel very welcome.’









No comments yet