Lipids have been the the poor relations of DNA and proteins for some time, but Philip Ball discovers that they are ready to take centre stage
When US biotechnology entrepreneur Craig Venter recently announced a $70 million (£42?million) project to understand ageing and longevity using genomics, Kai Simons was unimpressed. Genomics alone won’t get to grips with the body’s workings, he says. ‘We’ve learnt that the problem is very complex and you won’t get any mechanistic information by looking at all the genes that do this or that.’
A much more productive approach, Simons thinks, is buried away in the small print of Venter’s venture. ‘In addition to genome and microbiome data’, according to one report, the company will ‘collect data on biochemicals and lipids circulating throughout patients’ bodies’. Compared with genes, lipids – the small fatty molecules that constitute cell membranes, act as energy reserves, and mediate biochemical signalling – are deeply unglamorous. But many researchers agree with Simons, a biochemist at the Max Planck Institute for Molecular Cell Biology and Genetics in Dresden, Germany, that they might tell us much more about the body’s state of health.
This is why researchers worldwide are aiming to construct a complete picture of the body’s complement of lipid molecules: its lipidome.1 A US consortium called Lipid Maps, supported by the National Institutes of Health, has already been running since 2003 ‘to identify and quantitate all of the major, and many minor, lipid species in mammalian cells’. It aims to provide a ‘one-stop shop’ for lipid research. In the EU, the Seventh Framework Programme for Research funded LipidomicNet, in collaboration with Lipid Maps.
Lipids are very diverse – there might be thousands of different types in our bodies at any time, although probably only a much smaller number are informative about health and disease. But the real point about lipids is that they are closer to the surface of the action. Reading a genome might disclose predispositions to particular kinds of disease, and perhaps reveal individuals blessed with rather extreme longevity. But their message is much the same for a person as a newborn infant or as a septuagenarian, whereas lipids may reveal what the body has experienced, and how it has responded, in the intervening years. They are not genetically encoded, but are synthesised by enzymes that are themselves responsive to their environment – for example, to temperature or diet. In this way, lipids record what’s going on right now, just as the state of a nation is more sensitively judged by reading the papers than by consulting its dictionaries.
Lipids have already proved their value as a medical prognostic: witness the way doctors routinely check our cholesterol levels to gauge our vulnerability to heart disease. But reading the lipidome can also provide information about conditions such as diabetes, obesity and Alzheimer’s. What’s more, intervening in the body’s lipid chemistry could provide new avenues for drug development. And at the fundamental level it could help us to understand why cells have the shapes they have, how they communicate with one another, and how they store and use energy – in other words, why they are alive.
Taking shape
There is in a sense nothing new about the idea of lipidomics. In the 1960s, many biochemists were as interested in lipids as they were in DNA and proteins. But the latter took the prize – literally in the case of many Nobels. As it became ever more clear that genes and their interactions with proteins accounted for the most fundamental control mechanisms of living cells, lipids and other small-molecule constituents of cells such as hormones became sidelined as bit-part players, dancing to the tune of genes.
Yet much of the immediate ‘life of the cell’ hinges on lipids, and can’t be deduced at all at the genomic level. Even something as basic as the shape of a cell is largely controlled by its lipids; different cell types typically have distinctive shapes, so there is nothing arbitrary about it. Lipid membranes are also essential to the way cell components are moved around: proteins and other molecules are packaged up in lipid bubbles called vesicles, some of them ferried by motor proteins. For elucidating the mechanisms of this vesicle transport, James Rothman, Randy Schekman and Thomas Südhof were awarded the 2013 Nobel prize in medicine or physiology.
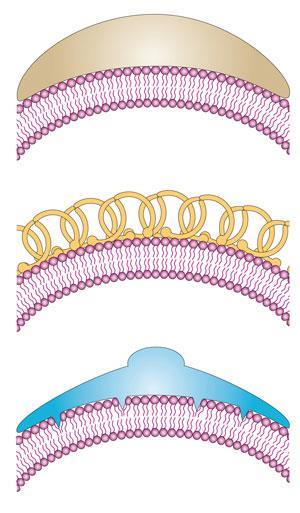
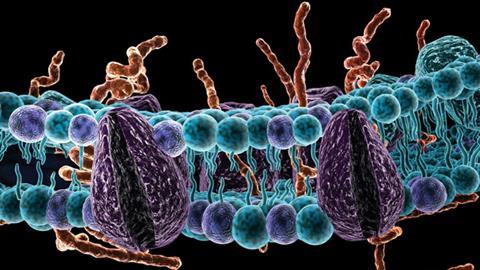
Lipids shape cells and their component organelles. They form membranes by aggregating into two-dimensional arrays – in effect liquid crystals – thanks to their Janus nature. The head groups are generally charged or polar groups that are water-soluble (hydrophilic), while the tails are insoluble (hydrophobic) fatty hydrocarbon chains or ring networks. Membrane lipids sit side-by-side in a ‘leaflet’ sheet, and two leaflets stick back-to-back in a bilayer with the hydrophobic tails on the inside, sheltering them from water.
Much of this was clear in the 1960s. The Italian biochemist Vittorio Luzzati showed that lipid bilayers in solution can form a variety of structures, including disorderly, labyrinthine ‘sponge’ phases that resemble the endoplasmic reticulum, the manufacturing and distribution hub of the cell. This network of folded lipid bilayer sheets surrounds the nuclei of eukaryotic cells, and is where proteins and other cell molecules (including lipids themselves) are synthesised by enzymes embedded in the membrane before being transported to their destination.
Lipid membranes were seen until quite recently as a passive fabric, with all the interesting stuff done by proteins. But there is an increasing understanding now that the shapes of membranes have a functional role. For example, the corkscrew-like shape of spirochaete bacteria might enable them to penetrate into host cells, while red blood cells can alter their shape very easily, assisting their hydrodynamic flow. Proteins actively control and alter the curvature of membranes,2 for example by binding to their surface as a rigid deforming ‘mould’ or by inserting fat-soluble appendages in between the lipids that act as curvature-inducing wedges. The budding of vesicles from membranes for cell traffic can involve a protein called clathrin that sticks to receptor molecules at the membrane surface and forms an enclosing protein coat. Not only does this mean that the lipid membranes are restructured, but that restructuring itself can encode information: some proteins can sense membrane curvature, so that the shape itself transmits a biochemical signal.3
More than walls
That’s not the only way in which lipid membranes can perform active signalling functions. Lipid membranes are composed of a variety of molecules, including phospholipids (which have phosphate–glyceride head groups), sphingolipids (with amino alcohol heads) and sterols like cholesterol. Some of these in turn have additional modifications such as sugar groups on their heads (glycolipids) which may mediate molecular recognition at the cell surface, for example determining blood groups. While the old view of lipid membranes regarded them as simple fluid mixtures of lipid types, it is now clear that different membranes have quite precisely tailored lipid compositions. The plasma membrane that constitutes the cell’s outer boundary, for instance, has mostly phospholipids (phosphatidylcholines) and sphingolipids on its outer leaflet, but the inner leaflet has rather different components. Sterols are more concentrated in the plasma membrane than in the endoplasmic reticulum. It evidently matters what goes where, but we don’t fully understand why.
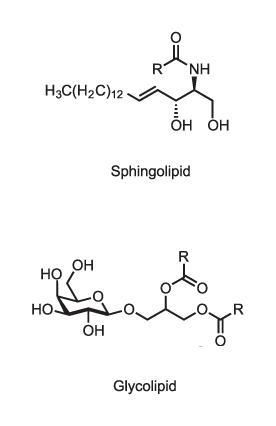
What’s more, within a given membrane different lipid types can be segregated into clumps, just like the separation of immiscible vinegar and oil in salad dressing. Sphingolipids and their glycosylated variants are particularly prone to this aggregation because they can link together via hydrogen bonds, either with each other or with the alcohol head group of cholesterol. Simons and his coworkers in Dresden have argued that the formation of these ‘lipid rafts’ has a functional role: that cells promote and stabilise it for specific signalling purposes.4 As a spontaneous phenomenon driven essentially by physical chemistry, it offers a very low-energy way to trigger physiological changes such as vesicle trafficking. Because particular membrane proteins might be more ‘soluble’ in some lipid patches than others (for example, the length of the proteins’ hydrophobic anchors should ideally match that of the surrounding lipids), lipid rafting might serve as a way of bringing different proteins together and assembling them into functional units.
In short, then, lipids in cell membranes can potentially encode and transmit signals in a variety of ways, drawing on their intrinsic chemical physics and their delicate and subtle interactions with proteins. But even that doesn’t exhaust the possibilities for lipids playing an active role in the conversations within cells. Some of them can act alone, rather than en masse, to trigger cell events and act as messengers to ferry information from one place to another.
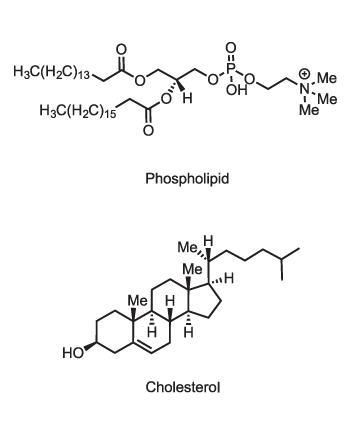
Although these various roles are known, there is still a great deal we don’t understand about what lipids do in the body. Even cholesterol itself is a mystery. ‘We don’t really understand what cholesterol is doing,’ says biochemist Teymuras Kurzchalia of the Max Planck Institute in Dresden. ‘Some say it makes membranes more fluid, some say just the opposite.’ But whatever it is doing in our own bodies, that role is clearly important, as it represents an immense energetic investment: about 50 molecules of ATP are needed to make one molecule of cholesterol. Evolution wouldn’t have bothered unless it was worth it.
Vital signs
Given all this business that lipids conduct, what might we expect to glean from monitoring our lipidome?
We know that lipids matter for human health – for example, the benefits of a diet rich in omega-3 fatty acids are well attested. But we don’t really know why, nor how lipid intake is related to lipid composition in the body. Lipidomics should change this. ‘It would not be surprising to find that many dietary superstitions vanished into thin air by accurate and biologically relevant measurements,’ say Simons and his colleague Andrej Shevchenko.5
Some researchers anticipate that lipidomics should improve our understanding of both the risks and the causes of a wide range of diseases, including diabetes, cardiovascular disease, disorders of the central nervous system, Alzheimer’s and some cancers. Where some of the lipid-based problems stem from lifestyle influences such as drinking, poor diet or smoking, as in heart disease, it might be possible to correct them with rather straightforward interventions. It is far easier to manipulate the lipidome than the genome.
Current approaches to monitoring lipid indicators, such as measuring HDL (‘good’) and LDL (‘bad’) cholesterol or triglycerides in blood serum, provide only a fraction of the information that a more comprehensive view of the lipid profile might offer. Peter Meikle of the Baker IDI Heart and Diabetes Institute in Melbourne, Australia, believes that an ability to examine perhaps 400 or so lipid species would greatly improve the risk predictions for these conditions. For example, by looking at 305 different lipids in patients, Meikle and colleagues were able to distinguish between those with clinically defined stable and unstable heart conditions.6
What’s more, lipid measurements might reveal a patient’s response to treatment. By looking at changes in lipid profiles following the use of statin drugs to regulate lipid production, it has proved possible to identify subjects for whom the drugs had little effect as well as possible biomarkers for their toxic side-effects. They also offer insights into the causes and consequences of weight loss for people with obesity. Although, as you’d expect, weight loss is a good thing, it’s not necessarily that simple: the problems with lipid metabolism, for instance, seem not to stem from obesity per se as much as from the specific daily caloric intake. It’s not just a matter of your weight but of what you’re eating.
Meikle’s group has also found that, by augmenting the usual indicators of risk of type 2 diabetes in the blood plasma of subjects (triglycerides and glycated haemoglobin, which reflects plasma glucose levels) with measurements of 21 lipid types in the blood plasma, they were able to improve the risk predictions significantly,7 correctly identifying an additional 20.7% of patients with type 2 diabetes or impaired glucose tolerance (albeit with an additional 2.4% of false positives).
Lipid drugs
If some lipids are functional molecules that trigger cell processes, we might reasonably expect them to be not only indicators of disease but potential targets for intervention. Again, however, proteins and DNA have dominated the scene here. There are already drugs that target enzymes related to lipid metabolism – most notably statins, which lower cholesterol levels by inhibiting the enzyme HMG-CoA reductase that is central to cholesterol manufacture in the liver. And Novartis’s Gilenya (fingolimod) is a recently approved drug that combats multiple sclerosis by mimicking the messenger lipid sphingosine 1-phosphate.
But could we also design drugs that bind to and modify the behaviour of lipids themselves? In principle, yes – and there are clear motives for doing so. For example, the cell membranes of tumour cells can have a different lipid composition from those of normal cells, containing more glycosphingolipids, so this could be exploited to target them. But pharmaceutical companies don’t like the membrane as a target, says Simons – not for any fundamental reason, but simply because pharma is a conservative business and they aren’t used to that strategy. ‘Lots of signalling receptors are modulated by glycolipids,’ says Simons, and so these could be good therapeutic targets. But because these lipids tend to function collectively, for example in lipid rafts, the drugs would operate differently: ‘It’s not going to be a lock and key principle,’ he says.

For one thing, it seems that it might not be a matter of simply making a molecule that attaches to the right lipid · antibodies that bind to glycosphingolipids seem to stick to two or more of them at a time. So the exact local composition and shape of a membrane might determine whether a drug targeting it can become attached. Nonetheless, some researchers are developing antibodies that can bind to lipids in mycobacteria such as M. tuberculosis and M. leprae (the agent of leprosy), both to identify and ultimately to combat the infections.
Understanding lipid metabolism – how these molecules are made in the body – could have some valuable spinoffs for chemicals manufacturing. Some lipids have important uses but are in short supply: the lubricant wax cetyl palmitate, for example, can no longer be legally obtained from its natural source in the heads of sperm whale, and a wax from the jojoba plant could be an industrial lubricant, rather than just a cosmetic ingredient, if it wasn’t so expensive. Ways to synthesise these substances artificially, perhaps by lipid metabolic engineering of bacteria, would be a welcome by-product of lipidomics.
Ultimately, though, Kurzchalia sees the challenge as that of understanding basic biology. You wouldn’t attempt to understand a material without knowing what it is made of, he says – and the same is true for the complex ‘soft matter’ that is a living organism. For that, he says, we need a ‘list of parts’ – and there’s no doubt that lipids are a vital component of those ingredients.
Yet if lipidomics is to succeed, says Simons, it needs to stake its claim: to make the intellectual case, but also to get young people interested in the field and train them in the right concepts · including the idea that life involves chemistry beyond genes and enzymes. That’s been a difficult task, he says, but he adds that ‘I think it will change in the next 10 years.’
Philip Ball is a science writer based in London, UK











No comments yet