A 2000 year old decapitated Yorkshire man and the ancient proteins in his preserved brain might provide clues to modern diseases, as Hayley Bennett discovers
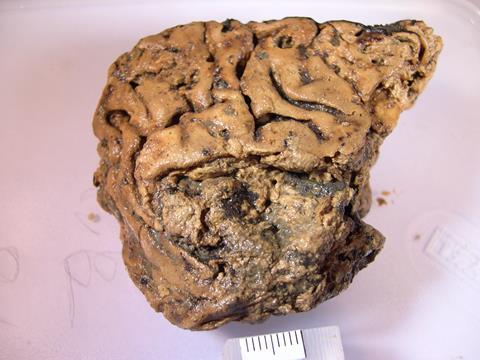
Neurologist Axel Petzold was listening to the radio, nursing a hangover from the previous night’s lab party, when he first heard about the Yorkshire man. It was December 2008 and, according to a news report, archaeologists had discovered the 2600-year-old skull of a decapitated Iron Age man buried in a wet pit at an excavation on the University of York campus in the UK. Inside it was something quite unexpected: his shrivelled, but very much intact brain.

The man apparently died in a brutal attack involving at least seven blows to the front of the neck. His brain, which should by all rights have decomposed long ago, was caked in mud but careful cleaning revealed its preserved structure, including the ridges and furrows that neurologists call gyri and sulci. ‘No one knew how this brain could survive,’ Petzold recalls. Immediately, though, he connected the remarkable finding to his own research. In his lab at University College London (UCL) Queen Square Institute of Neurology, Petzold was trying to help patients suffering from vision loss caused by damaged nerves in their brains. This involved looking for certain proteins that populate nerve cells. It struck him that some of the peculiarities of these proteins might account for the exceptional preservation of the Yorkshire man’s brain.
Captivated by the radio report, Petzold was quick to get in touch with the York group. Even as his hangover was receding, he found himself on the phone to lead archaeologist, Sonia O’Connor. ‘By then I had managed to get an aspirin and get back to work,’ he laughs. ‘And we had a very nice discussion.’
Palaeo-curious
With Petzold’s help, over the next decade, the team began unravelling the beheaded man’s story via the proteins in his shrunken brain. To do it, they used biochemical techniques once more familiar to molecular biologists, but that are now part of the toolkit of those involved in ‘palaeoproteomics’ – the study of ancient proteins. While Petzold was by no means a palaeoproteomics expert, he was what you might call palaeo-curious, having shared the queue for his hospital’s CT scanner with mummies from the British Museum, just over the road. He was also well-acquainted with the molecular constituents of the human brain.
In Petzold’s own toolkit were antibody-based tests he used to pick floppy protein fibres called intermediate filaments from the brains of living patients. Now, he intended to apply similar tests to the long-dead. It was these fibres that Petzold suspected had a part to play in preserving the Iron Age brain. Intermediate filaments are 8–12nm thick proteins that give structural stability to brain cells, while also being flexible enough to change configuration. They take two forms: neurofilaments (in nerve cells) and glial fibrillary acidic proteins (GFAP) (in supporting cells called astrocytes). Neurofilaments are useful biomarkers for nerve cell damage because they leak from injured neurons; Petzold was measuring levels in patients with traumatic brain injuries using a probe called a microdialysis catheter inserted into the frontal cortex, a way of monitoring brain biochemistry ‘live’. ‘You can have it sitting directly next to the neurons in the brain,’ he explains. ‘If we use that type of fluid, then you can still see the full-length protein.’
In the Yorkshire man’s brain, by contrast, these proteins were far from whole. But they were still there – as Petzold’s test proved – even if they had been slowly fragmenting for thousands of years. It’s not unheard of for proteins to survive this long. Proteins degrade far more slowly than DNA and are increasingly giving us information about the deeper past. Animal bones, which contain collagen proteins that can be matched to certain species, are common sources, for example, allowing archaeologists to illuminate animal husbandry practices of the past.
Meanwhile, scrapings from human teeth have helped Jessica Hendy, a palaeoproteomics expert working separately from O’Connor at the University of York, to uncover direct evidence of our ancestors’ diets. ‘We did some protein extractions on dental tartar and one of the things that kept jumping out to us was evidence of milk proteins,’ Hendy says. ‘So we’ve discovered this as a new way to identify milk consumption in the past.’ She adds that the history of humans’ milk habit is particularly fascinating for archaeologists because it’s only relatively recently we evolved to digest lactose. By looking for differences in the amino acid building blocks of ancient milk proteins, it’s also possible tell whether they came from cows, sheep or goats.
More brains
What’s more surprising than getting proteins from old bones or teeth, however, is getting them from an old brain, a soft tissue that we assume would be ravaged away relatively quickly by the normal processes of putrefaction. Alexandra Morton-Hayward, an ex-undertaker now studying the biomolecular composition of preserved brains for her doctoral degree at the University of Oxford, UK, has been discovering that brains don’t always decompose as expected. The Yorkshire man was her way into the subject too – she started digging into the literature when she came across a paper about him. ‘One reference led to another and to several more,’ she recalls. Before she knew it she had found reports of more than 1800 preserved brains published since the mid-1600s. Although she doesn’t have one coherent explanation for why all these brains survived – they occur in dry and wet conditions and in different soils – she too is interested in the floppy protein components.
As well as the intermediate filaments, Morton-Hayward is looking at myelin basic protein (MBP), a key central nervous system protein that was also recovered from the Yorkshire man. All three are shapeshifters. They have ‘intrinsically disordered regions’ enabling them to refold like origami paper instead of forming stable 3D structures. As Morton-Hayward explains, their flexible structure is closely tied to their function in the ‘constant re-routing and rewiring’ of ‘mind-blowingly elaborate motor pathways’, or neuroplasticity – the brain’s way of adapting to experience, and to damage.
Like other researchers, Morton-Hayward’s go-to tool for detecting these proteins is mass spectrometry, which she is currently using with material extracted from the brains of hundreds of people buried in a 19th century British workhouse cemetery. As in modern protein studies, the basic premise is this: extract proteins, digest using enzymes, then smash them up in a mass spectrometer and use software to look at the pattern or spectrum that the pieces produce and make matches to reference proteins. It was the next obvious tool in the kit for Petzold too.
Using mass spec, it’s now possible to pull sequences for fragmented proteins from within the chemical milieu of ancient tissues and also to explore how the protein contents vary for each tissue – one of the benefits of working with the proteome rather than the genome. Experts get a very different set of molecules for the brain compared to bone, which is mostly dominated by collagen, or for, say, faeces (see box Poop proteome below).
Poop proteome
Like brains, coprolites (preserved faeces) tend to be under-recognised and under-reported – possibly, as far as the poop is concerned, due to some squeamishness on the part of researchers. Recently, however, Hendy’s team used mass spec to analyse proteins from 300-year-old (or so) sled dog faeces from an indigenous Alaskan community. The deposits, preserved by the frigid conditions, ‘still smelt like poop’, she says. They were able to tell what kind of fish the dogs ate by analysing small fragments of fishbone within the excrement using a quick, cheap mass spec technique called ‘peptide mass fingerprinting’, which generates a ‘barcode’ for a species based on the particular pattern its collagen components produce.
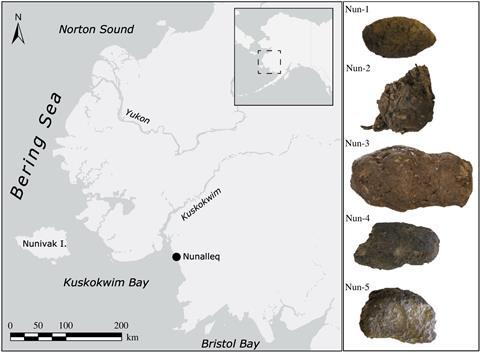
To drill down into the poop proteome itself, however, they used liquid chromatography tandem mass spectrometry (LC–MS/MS), which separates out the protein fragments by chromatography before firing them at the collector. Via this route, they identified 56 different dog and fish proteins in the ancient faeces, from digestive proteins to proteins from fish guts, muscle and roe. These findings led to insights that wouldn’t be possible with DNA: for example, that the faeces were probably from the summer, when the dogs were fed the guts and roe of fish caught and gutted during the seasonal salmon run.
Protein problem
What can you expect to find in a human brain, except, well, brain proteins? In 2013, a European team used LC-MS to study tissue samples from the brain of Ötzi the iceman, the well-preserved body of a man who died 5300 years ago in the Alps, pulling out more than 500 separate proteins. Among them were some related to stress and bleeding that led the researchers to come up with a new theory for how Ötzi died – from a head injury. Of course, the Yorkshire man’s mode of death wasn’t in question but, nevertheless, the mass spec results were fascinating: 783 protein matches in total, a triumph for such an ancient soft tissue. ‘They actually managed to recover the richest palaeoproteome from any archaeological material investigated to date,’ Morton-Hayward notes. Among the molecules they identified were the three floppy proteins again: neurofilaments, GFAP and MBP.

Progress generally, though, was fairly slow. As a clinician, Petzold was working on the project in his spare time and he kept coming up against strange results. Firstly, his extractions from the Yorkshire man’s tissue contained an impenetrable brown muck that he suspected was protein but which couldn’t be accessed in tests. Meanwhile, in some tissue samples he’d left to decay for a year, he was seeing the opposite of what he’d expected – instead of protein levels dropping off as the tissue decayed, they were increasing. So on the one hand there was missing protein and on the other hand, protein appearing where it shouldn’t be.
Intriguingly, Morton-Hayward says there’s also something missing from the data on her Victorian brains. The mass spec analysis is only matching about half of the MBP protein sequence; the N-terminal half is ‘basically missing’ from her samples, despite them being much younger than Petzold’s. It’s as though, she says, her methods ‘aren’t accessing all of the protein that’s there’. She also points out that proteins like MBP and the floppy intermediate filaments have a propensity to aggregate and form plaques, like the ones in the various forms of dementia.
The brain shrinks and shrinks but these proteins still preserve the architecture of the cells
This aggregation, it seems, could be the solution to both mysteries. Petzold eventually concluded that his samples were full of aggregated proteins, which were slowly unravelling themselves. So every time he used his antibody tests on the decaying samples, he detected more than before, as more latching-on spots for his antibodies were revealed. The brown muck, he reasoned, must also contain aggregated proteins, explaining why he couldn’t access it. In 2020, Petzold finally published a theorv explaining what kept the beheaded man’s brain from disintegrating. It’s the first work to suggest that protein aggregation could be helping to preserve ancient brain tissue. ‘That’s the trick of these proteins,’ Petzold says. ‘The brain shrinks and shrinks but these proteins still preserve the architecture, the structure of the cells.’
Morton-Hayward says she’s keen to see if she can replicate this finding of proteins aggregating and disaggregating, perhaps initially with modern brain tissue. But if aggregation did have a part to play in both the Victorian brains and the Yorkshire man’s, then why aren’t all brains preserved this way? All, presumably, contain the necessary molecular constituents. One theory might centre on the influence of neurodegenerative conditions, like dementia, linked to protein aggregation and plaque formation. In the case of the workhouse, such disorders may have been more prevalent due to deprivation.
According to Morton-Hayward, the aggregation process is spontaneous – not driven by enzymes – and might even continue after death, so that over hundreds or thousands of years some really stubborn plaques could form, the like of which couldn’t be observed even in the oldest living centenarians. ‘That’s something I find really exciting,’ she says, hinting that a load of long-dead brains could actually deepen our understanding of the molecular pathways involved in certain neurological diseases.
Bioinformatics bonanza
As for the ill-fated Yorkshire man, Petzold already has plans in place for a more thorough investigation of his brain proteome – looking specifically for the molecular hallmarks of neurodegenerative diseases. He believes some of the signs are in the mass spec data they already have, but due to the high level of statistical rigour in their analysis, some of the protein sequences they originally identified had to be discarded. As Morton-Hayward notes, it’s a case of selecting appropriate thresholds for pattern-matching your proteins with the reference proteins provided by the software: ‘a game of snap’ but with a confidence interval. Whether you say snap depends on how closely you decide the cards have to match. If you set your threshold for saying snap too low, then you can deal a protein sequence and match it to the wrong reference sequence, whereas in Petzold’s case, he wonders whether the bar was set too high.
There is more to the data processing than just threshold-setting, though. As Frido Welker at the University of Copenhagen in Denmark explains, it’s advances on the bioinformatics side – rather than on the protein extraction side – that have really driven palaeoproteomics forward in the last few years. ‘I think that aspect is actually core to retrieving more protein sequences, being able to reconstruct the sequences at higher coverage and higher reliability, but also then to finding more proteins,’ he says.
It’s a game of snap but with a confidence interval
In 2020, Welker’s team published proteome data from two incredibly old dental enamel samples taken from ancient hominins. Here, their protein search strategy was crucial to dealing with very degraded proteins, which were 800,000 years old for Homo antecessor, a potential common ancestor of humans and Neanderthals, and 1.8 million years old for Homo erectus. In such old samples, the passage of time allows chemical changes to accumulate in the side chains of amino acids within the proteins. These changes are actually used in palaeoproteomics to verify that the proteins are genuine relics and not just the result of contamination from modern sources. However, they also cause what are known as mass shifts – differences between the mass spectra obtained from ancient and modern proteins. For Welker, this meant building in a level of error-tolerance to account for differences in the very old protein sequences he was detecting. Such changes, termed post-translational modifications or PTMs, were present and had to be accounted for in the Yorkshire man’s proteins too.
Morton-Hayward also has a keen interest in optimising these ‘downstream’ aspects of protein identification. It is possible, she explains, to increase protein recovery by making minor tweaks to the software settings even once the data has been collected and uploaded. For example, giving more information about how the protein was digested and factoring in the ‘digestion’ caused by time itself tells the software to look for different kinds of fragments. ‘It massively increases your search space,’ she says.
It’s the software side of things that Petzold says he needs to get a handle on if they’re ever going to truly solve the case of the beheaded Yorkshire man. Did some sort of neurodegenerative disorder influence the protein aggregation in his brain? Could the effects of this disease have led to the proteins and tissue being preserved for 2600 years? Only more time – and, likely, some more spectacular mass spec results – will tell. The alternative explanation is rather grislier. Based on the distribution of preserved proteins in the man’s brain, and corrosion of the knife used to dissect it, it seems plausible that his head could have been dropped into a barrel of acid soon after death – another excellent method for preserving proteins.
Hayley Bennett is a science writer based in Bristol, UK












No comments yet