Breakthrough technique allows specific cell proteins to be visualised at high resolution using EM
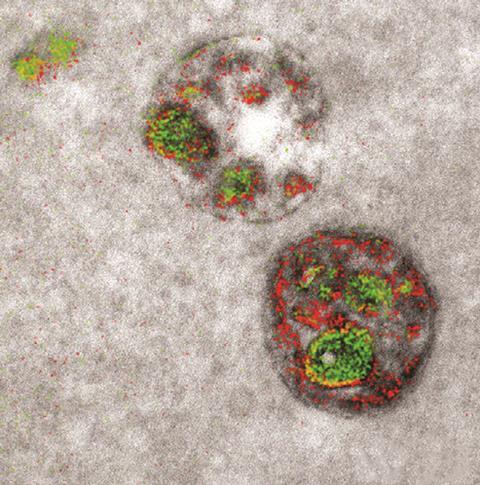
The greys of electron microscope images have acquired an injection of colour for the first time thanks to researchers in the US.
It is something that the labs of Mark Ellisman and Roger Tsien at the University of California, San Diego have been working on for 15 years. Tsien – who died unexpectedly earlier this year – was awarded the Nobel Prize in 2008 for the discovery of Green Fluorescent Protein (GFP), which is widely used in cell imaging.
‘This is the electron microscope extension of the revolution that happened with green flourescent protein, which has propelled biology so dramatically over the last 2 decades,’ says Ellisman.
Until now, visualisation of specific proteins in electron microscopy could only be achieved using antibodies conjugated to gold particles or quantum dots, but poor penetration into cells or tissues limited their usefulness. For the new technique, photosensitive proteins are encoded in the DNA of a cell. When these proteins are hit by light they spew out reactive oxygens.
‘The work represents an important breakthrough,’ comments chemist Alice Ting at Stanford University, US, who was not involved in the research. Though electron microscopy has much higher spatial resolution than fluorescence microscopy, it has only one ‘colour’ - black, which represents objects or regions in the sample that do not transmit electrons. ‘This work addresses [this drawback], by using clever lanthanide ligand chemistry in combination with [electron energy loss spectroscopy] to essentially perform ‘multicolour’ electron microscopy,’ Ting says.
The technique itself is complicated, involving a large number of extra specimen preparation steps. However, the authors are confident it can be moved from their custom-built microscope to a more standard issue. ‘And then we will work on how to make this multiplex to even more colours,’ says Ellisman.
Chain reaction
The cells are grown and then bathed in diaminobenzidine (DAB) monomers that contain a rare earth such as cerium or lanthanum. The small monomers easily diffuse across the cell. Then when photosensitive proteins are hit with a certain wavelength of light, they release reactive oxygen species. Any DAB monomer within 5nm is hit by these reactive species, causing it to polymerise.
‘We get a huge amplification, because it is like a chain reaction. The diaminobenzidine polymerises in one spot in response to the light,’ says Stephen Adams, another member of the research team. ‘The metal remains bound tightly to it via its chelator.’ The excess DAB monomer is then washed out of the specimen, leaving the polymerised DAB containing the rare earth element tightly bound in the specimen.
The specimen can then be infused with another flush of DAB monomers containing a second rare-earth metal, such as prasesodymium, and the process repeated. This is used to label a second protein with the help of a different photosensitizer that also generates reactive oxygen, though at a different wavelength of light. The distribution of the rare earths bound to the small region of poylmerised DAB is mapped using energy-loss spectra, which is distinctive for each element.
‘The electrons interact with the sample and a fraction of them lose a few hundred volts by interacting with lathanide atoms,’ Ellisman explains. ‘This is used a lot in materials science, looking for the elemental composition of solids,’ adds Adams. ‘You take a greyscale image first, with a conventional electron microscope, and add a filter to pick out these specific absorbances.’ Reds and greens are used to mark the location of the lathanides on the otherwise grey image.
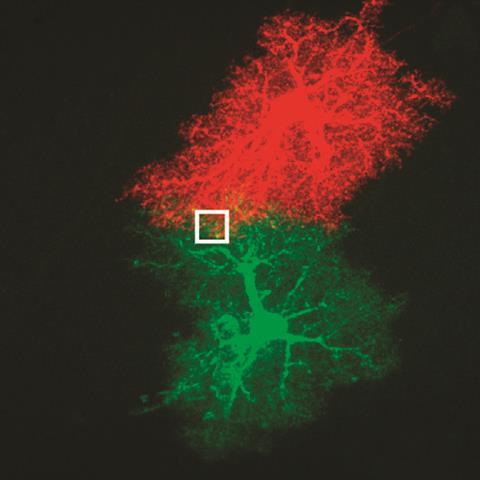
The technique was used to track polyarginine peptides as they enter a cell, which Tsien’s lab was using to detect tumour cells. It was also used to track the interactions of star-shaped nerve cells called astrocytes. And it showed that newly synthesised protein kinase M zeta in cultured neurons preferentially travels to the post-synaptic membrane.
‘This is another project from Roger Tsien’s lab looking at where long-term memories are stored in the brain. We wanted to see if these newly made molecules were located in a part of the synapses,’ Adams explains. ‘We are now working on three colours so that we can visualize three components in the brain of a mouse at super-high resolution.’ This chases up a hypothesis Tsien proposed in 2013 on how long-term memories are stored in the brain. Ellisman is now looking at the fundamental structure of our genome. ‘We want to see how the nucleus is organised and which proteins are doing what,’ he says.
References
S R Adams et al, Cell Chemical Biology, 2016,23, 1, (DOI: 10.1016/j.chembiol.2016.10.006)








No comments yet