New method uses gold nanoprobes to overcome the problems of PCR detecting small nucleic acids
The polymerase chain reaction (PCR) is the way to go if you want to detect small amounts of nucleic acids like DNA. But this technique requires expensive reagents, complex instruments and a repetitive, sometimes error-prone amplification process. Now, Korean chemists have come up with a new way to detect ultra-small amounts of DNA. They call it associating and dissociating nanodimer analysis or, simply, ADNA.
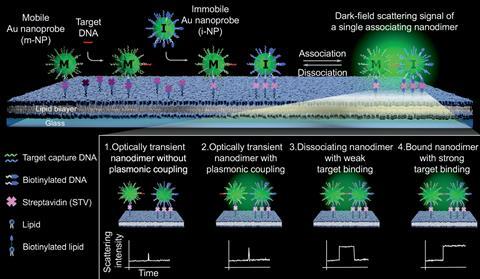
The technique uses gold nanoparticles covered with DNA sequences complementary to the ones they want to detect. Some are immobilised into a lipid bilayer – others remain free in solution – and their surfaces are observed with dark-field microscopy. ‘Dark-field microscopy is particularly useful to image and analyse metal nanoparticles that generate strong scattering signals and unique colours based on particle size, shape and composition,’ explains Jwa-Min Nam, lead author and researcher at Seoul National University in South Korea. ‘Moreover, the ‘dark-field microscopic setup is very simple, easy to use and inexpensive,’ he adds.
Thanks to the complementarity of DNA bases, when a mobile nanoparticle finds a piece of target DNA it immediately binds to it. Soon, it finds one of the immobilised gold nanoprobes and joins to it. This interaction between the nanoparticles changes the way they scatter light and this can be detected and analysed in real-time.
Nils Walter, a single molecule analysis researcher at the Univeristy of Michigan, US, and co-founder of Alight Sciences, believes ‘this paper provides a technology that works reliably, sensitively, avoiding the need of complicated workups of the samples, making analyses easier to perform’. Like the authors, he says direct detection methods compare favourably with PCR. ‘[PCR] works on assumptions that all biomarkers can be equally detected, and needs unreliable enzymatic amplification […] among other issues. Direct detections overcome these challenges, which is one of the steps forward of this work.’ Walter also finds important that the authors show ‘a proof of principle with human serum’, which is quite close to a real-life sample.
ADNA can reach the detection limits of PCR and go even further. ‘[Our] technique also allows the quantification of short oligonucleotides such as microRNA, which are difficult to detect with PCR due to the difficulty of designing the proper primer sequences,’ explains Nam. ‘It also minimises false positive signals, allowing reliable quantification of target DNA sequences.’ Walter adds: ‘This technique discriminates against spurious background binding events, […] it has a lot of promise.’ And applications for this type of technology could be on the horizon. ‘We just created a biotech start-up to commercialise a similar approach!’ concludes Walter.
References
K Kim et al., Angew. Chem. Int. Ed., 2017, DOI: 10.1002/anie.201705330















No comments yet